A family of unconventional deubiquitinases with modular chain specificity determinants.
Hermanns, T., Pichlo, C., Woiwode, I., Klopffleisch, K., Witting, K.F., Ovaa, H., Baumann, U., Hofmann, K.(2018) Nat Commun 9: 799-799
- PubMed: 29476094
- DOI: https://doi.org/10.1038/s41467-018-03148-5
- Primary Citation of Related Structures:
6EI1 - PubMed Abstract:
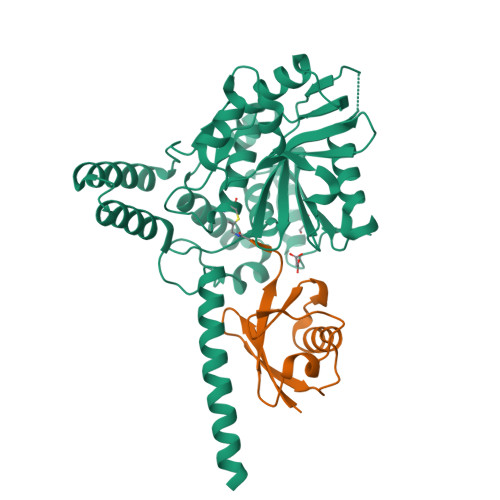
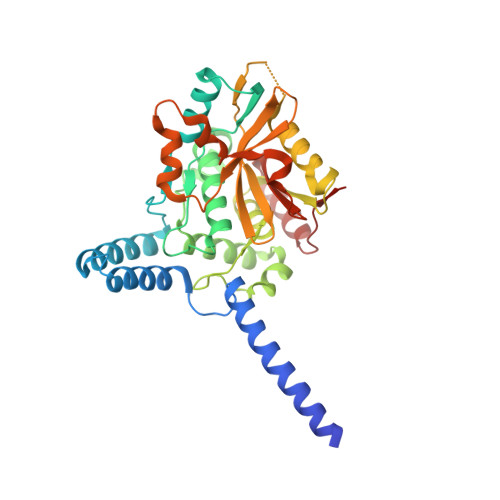
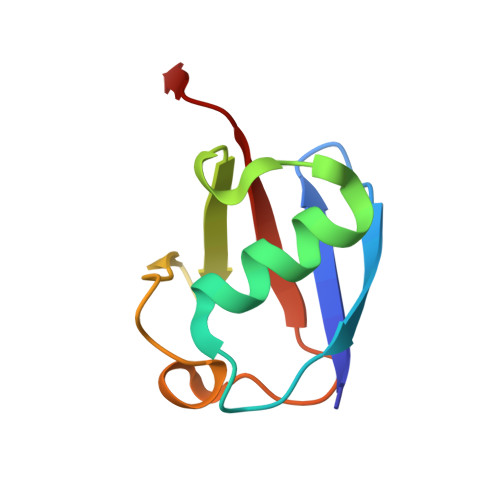
Deubiquitinating enzymes (DUBs) regulate ubiquitin signaling by trimming ubiquitin chains or removing ubiquitin from modified substrates. Similar activities exist for ubiquitin-related modifiers, although the enzymes involved are usually not related. Here, we report human ZUFSP (also known as ZUP1 and C6orf113) and fission yeast Mug105 as founding members of a DUB family different from the six known DUB classes. The crystal structure of human ZUFSP in covalent complex with propargylated ubiquitin shows that the DUB family shares a fold with UFM1- and Atg8-specific proteases, but uses a different active site more similar to canonical DUB enzymes. ZUFSP family members differ widely in linkage specificity through differential use of modular ubiquitin-binding domains (UBDs). While the minimalistic Mug105 prefers K48 chains, ZUFSP uses multiple UBDs for its K63-specific endo-DUB activity. K63 specificity, localization, and protein interaction network suggest a role for ZUFSP in DNA damage response.
Organizational Affiliation:
Institute for Genetics, University of Cologne, Zülpicher Str. 47a, 50674, Cologne, Germany.