A Triazole Disulfide Compound Increases the Affinity of Hemoglobin for Oxygen and Reduces the Sickling of Human Sickle Cells.
Nakagawa, A., Ferrari, M., Schleifer, G., Cooper, M.K., Liu, C., Yu, B., Berra, L., Klings, E.S., Safo, R.S., Chen, Q., Musayev, F.N., Safo, M.K., Abdulmalik, O., Bloch, D.B., Zapol, W.M.(2018) Mol Pharm 15: 1954-1963
- PubMed: 29634905
- DOI: https://doi.org/10.1021/acs.molpharmaceut.8b00108
- Primary Citation of Related Structures:
6BWP, 6BWU - PubMed Abstract:
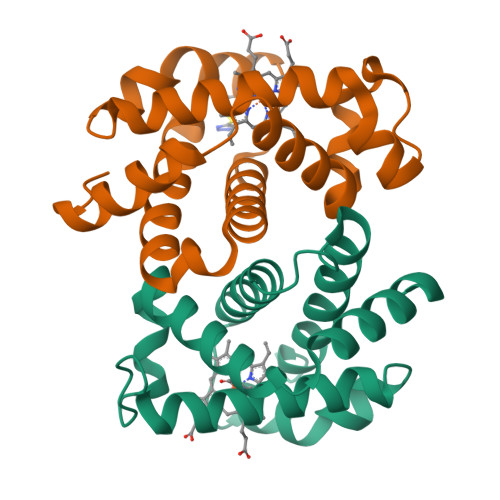
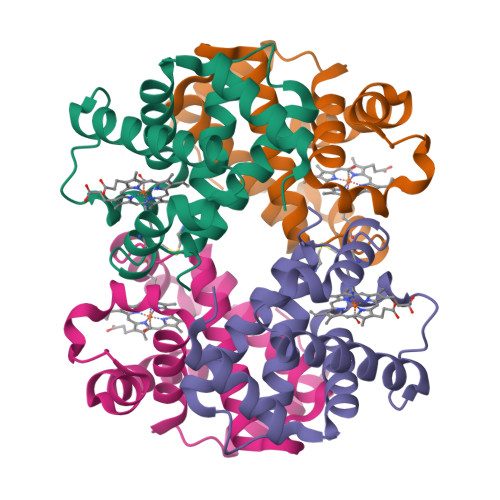
Sickle cell disease is an inherited disorder of hemoglobin (Hb). During a sickle cell crisis, deoxygenated sickle hemoglobin (deoxyHbS) polymerizes to form fibers in red blood cells (RBCs), causing the cells to adopt "sickled" shapes. Using small molecules to increase the affinity of Hb for oxygen is a potential approach to treating sickle cell disease, because oxygenated Hb interferes with the polymerization of deoxyHbS. We have identified a triazole disulfide compound (4,4'-di(1,2,3-triazolyl)disulfide, designated TD-3), which increases the affinity of Hb for oxygen. The crystal structures of carboxy- and deoxy-forms of human adult Hb (HbA), each complexed with TD-3, revealed that one molecule of the monomeric thiol form of TD-3 (5-mercapto-1H-1,2,3-triazole, designated MT-3) forms a disulfide bond with β-Cys93, which inhibits the salt-bridge formation between β-Asp94 and β-His146. This inhibition of salt bridge formation stabilizes the R-state and destabilizes the T-state of Hb, resulting in reduced magnitude of the Bohr effect and increased affinity of Hb for oxygen. Intravenous administration of TD-3 (100 mg/kg) to C57BL/6 mice increased the affinity of murine Hb for oxygen, and the mice did not appear to be adversely affected by the drug. TD-3 reduced in vitro hypoxia-induced sickling of human sickle RBCs. The percentage of sickled RBCs and the P 50 of human SS RBCs by TD-3 were inversely correlated with the fraction of Hb modified by TD-3. Our study shows that TD-3, and possibly other triazole disulfide compounds that bind to Hb β-Cys93, may provide new treatment options for patients with sickle cell disease.
Organizational Affiliation:
Anesthesia Center for Critical Care Research, Department of Anesthesia, Critical Care, and Pain Medicine , Massachusetts General Hospital and Harvard Medical School , Boston , Massachusetts 02114 , United States.