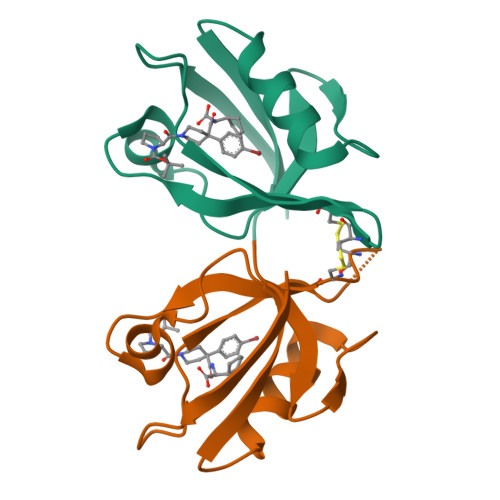
Lock and chop: A novel method for the generation of a PICK1 PDZ domain and piperidine-based inhibitor co-crystal structure.
Marcotte, D.J., Hus, J.C., Banos, C.C., Wildes, C., Arduini, R., Bergeron, C., Hession, C.A., Baker, D.P., Lin, E., Guckian, K.M., Dunah, A.W., Silvian, L.F.(2018) Protein Sci 27: 672-680
- PubMed: 29280296
- DOI: https://doi.org/10.1002/pro.3361
- Primary Citation of Related Structures:
6BJN, 6BJO - PubMed Abstract:
The membrane protein interacting with kinase C1 (PICK1) plays a trafficking role in the internalization of neuron receptors such as the amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) receptor. Reduction of surface AMPA type receptors on neurons reduces synaptic communication leading to cognitive impairment in progressive neurodegenerative diseases such as Alzheimer disease. The internalization of AMPA receptors is mediated by the PDZ domain of PICK1 which binds to the GluA2 subunit of AMPA receptors and targets the receptor for internalization through endocytosis, reducing synaptic communication. We planned to block the PICK1-GluA2 protein-protein interaction with a small molecule inhibitor to stabilize surface AMPA receptors as a therapeutic possibility for neurodegenerative diseases. Using a fluorescence polarization assay, we identified compound BIO124 as a modest inhibitor of the PICK1-GluA2 interaction. We further tried to improve the binding affinity of BIO124 using structure-aided drug design but were unsuccessful in producing a co-crystal structure using previously reported crystallography methods for PICK1. Here, we present a novel method through which we generated a co-crystal structure of the PDZ domain of PICK1 bound to BIO124.
Organizational Affiliation:
Biotherapeutics and Medicinal Sciences, Biogen Inc, Cambridge, Massachusetts.