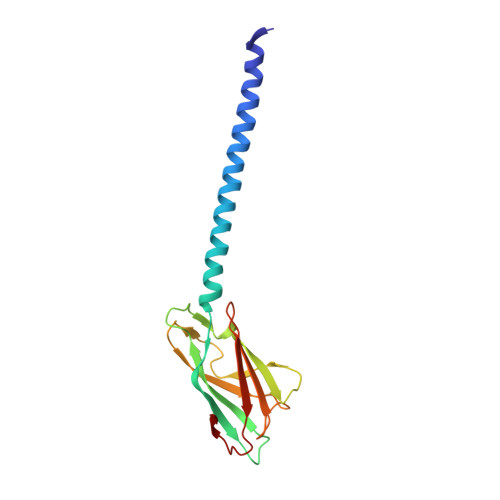
CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pili.
Poweleit, N., Ge, P., Nguyen, H.H., Loo, R.R., Gunsalus, R.P., Zhou, Z.H.(2016) Nat Microbiol 2: 16222-16222
- PubMed: 27922015
- DOI: https://doi.org/10.1038/nmicrobiol.2016.222
- Primary Citation of Related Structures:
5TFY - PubMed Abstract:
Archaea use flagella known as archaella-distinct both in protein composition and structure from bacterial flagella-to drive cell motility, but the structural basis of this function is unknown. Here, we report an atomic model of the archaella, based on the cryo electron microscopy (cryoEM) structure of the Methanospirillum hungatei archaellum at 3.4 Å resolution. Each archaellum contains ∼61,500 archaellin subunits organized into a curved helix with a diameter of 10 nm and average length of 10,000 nm. The tadpole-shaped archaellin monomer has two domains, a β-barrel domain and a long, mildly kinked α-helix tail. Our structure reveals multiple post-translational modifications to the archaella, including six O-linked glycans and an unusual N-linked modification. The extensive interactions among neighbouring archaellins explain how the long but thin archaellum maintains the structural integrity required for motility-driving rotation. These extensive inter-subunit interactions and the absence of a central pore in the archaellum distinguish it from both the bacterial flagellum and type IV pili.
- Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles (UCLA), Los Angeles, California 90095, USA.
Organizational Affiliation: