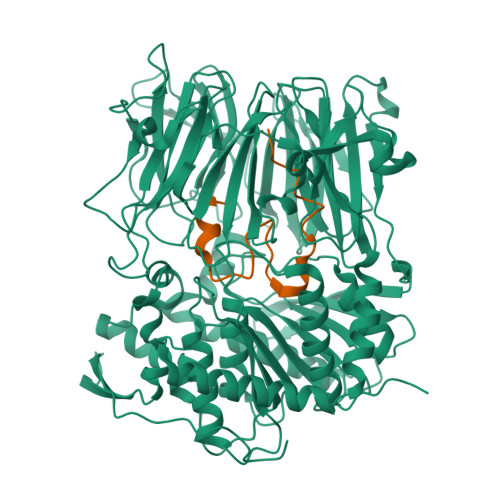
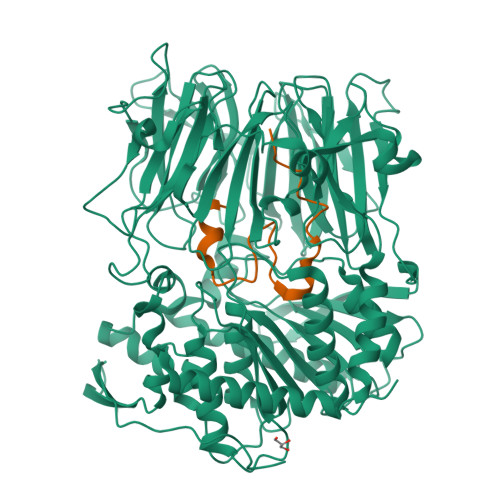
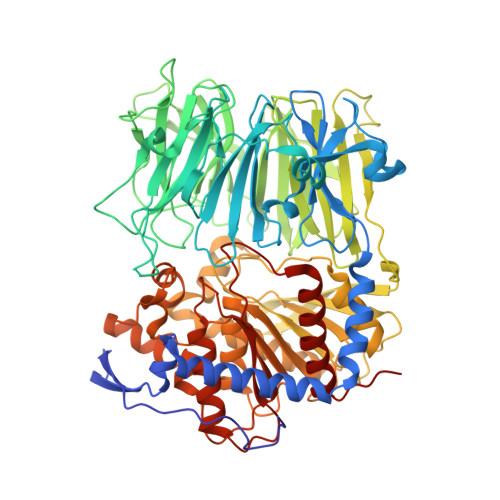
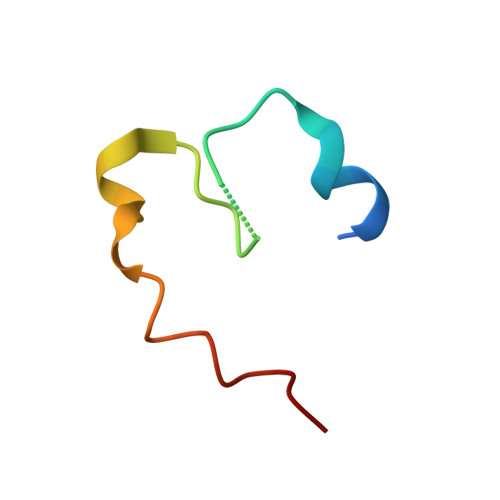
Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Czekster, C.M., Ludewig, H., McMahon, S.A., Naismith, J.H.(2017) Nat Commun 8: 1045-1045
- PubMed: 29051530
- DOI: https://doi.org/10.1038/s41467-017-00862-4
- Primary Citation of Related Structures:
5N4B, 5N4C, 5N4D, 5N4E, 5N4F - PubMed Abstract:
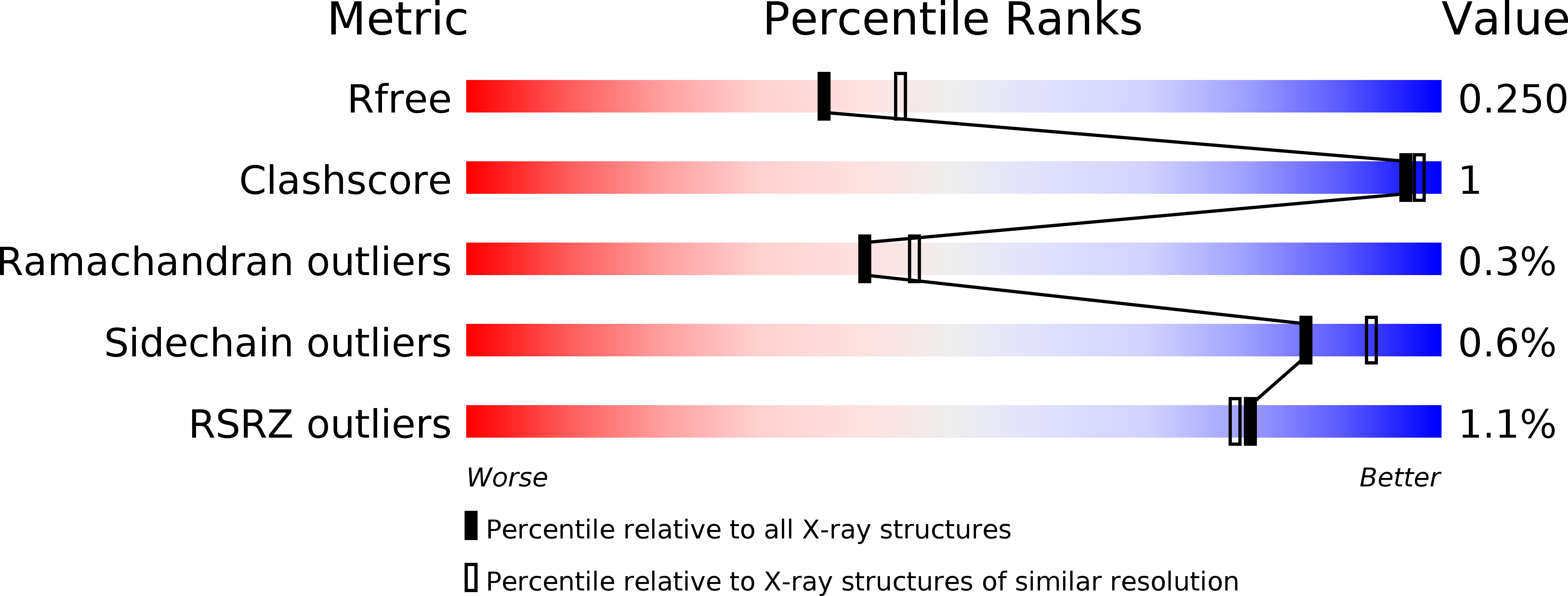
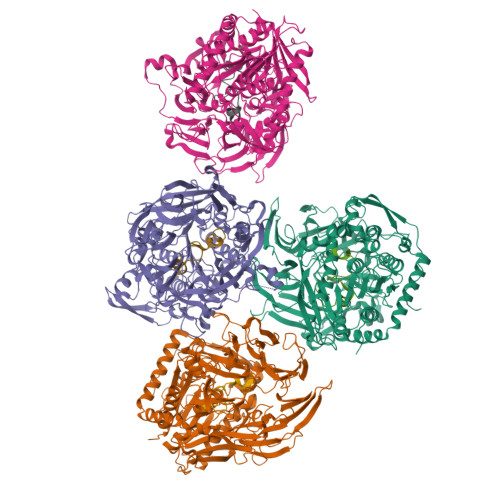
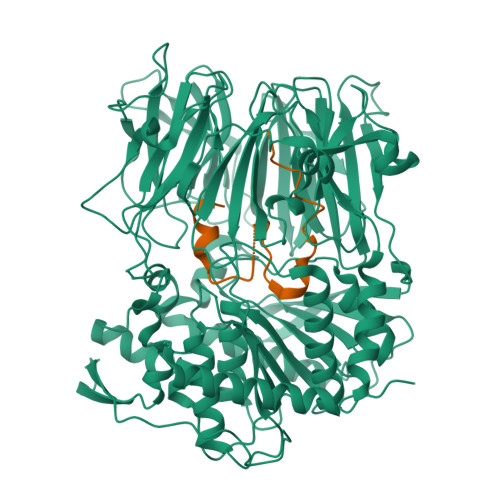
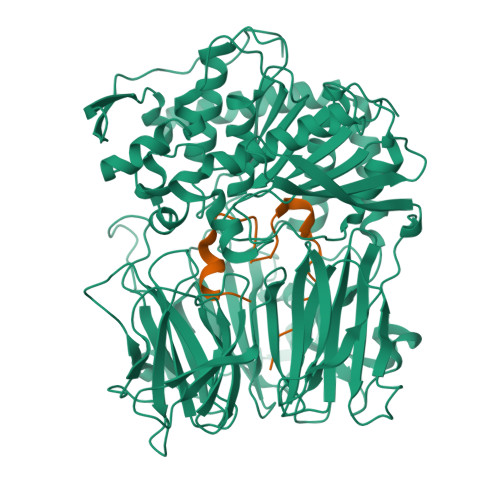
Peptide macrocycles are promising therapeutic molecules because they are protease resistant, structurally rigid, membrane permeable, and capable of modulating protein-protein interactions. Here, we report the characterization of the dual function macrocyclase-peptidase enzyme involved in the biosynthesis of the highly toxic amanitin toxin family of macrocycles. The enzyme first removes 10 residues from the N-terminus of a 35-residue substrate. Conformational trapping of the 25 amino-acid peptide forces the enzyme to release this intermediate rather than proceed to macrocyclization. The enzyme rebinds the 25 amino-acid peptide in a different conformation and catalyzes macrocyclization of the N-terminal eight residues. Structures of the enzyme bound to both substrates and biophysical analysis characterize the different binding modes rationalizing the mechanism. Using these insights simpler substrates with only five C-terminal residues were designed, allowing the enzyme to be more effectively exploited in biotechnology.
Organizational Affiliation:
Biomedical Sciences Research Complex, The University of St Andrews, North Haugh, St Andrews, K16 9ST, UK.