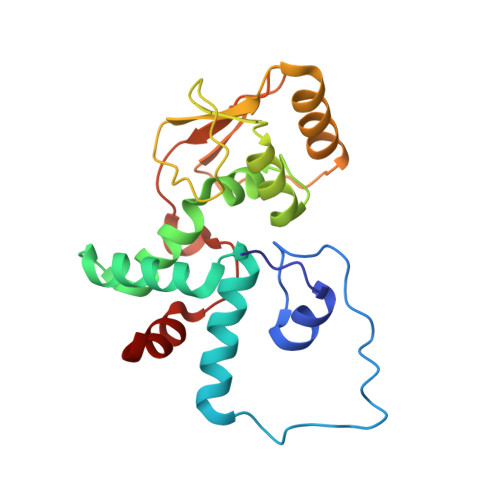
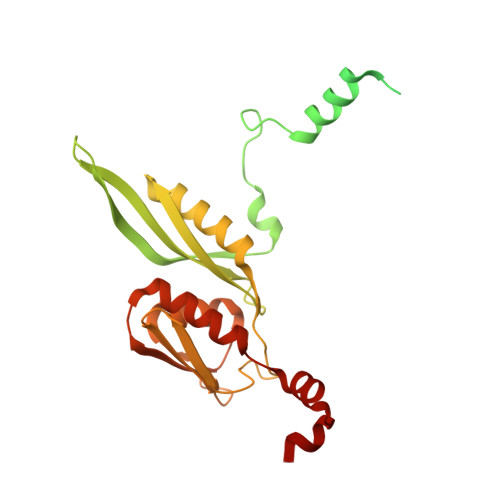
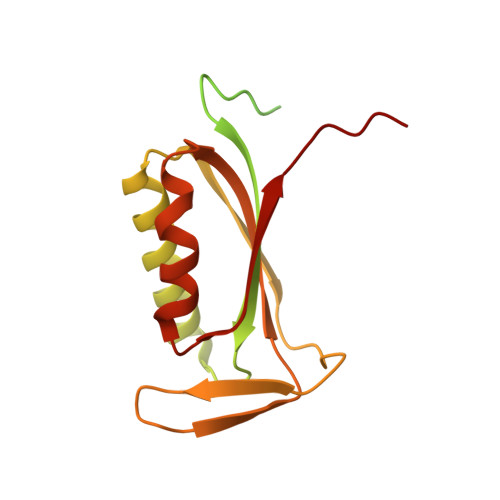
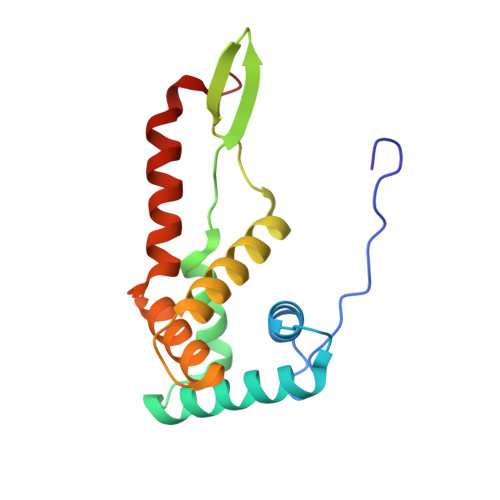
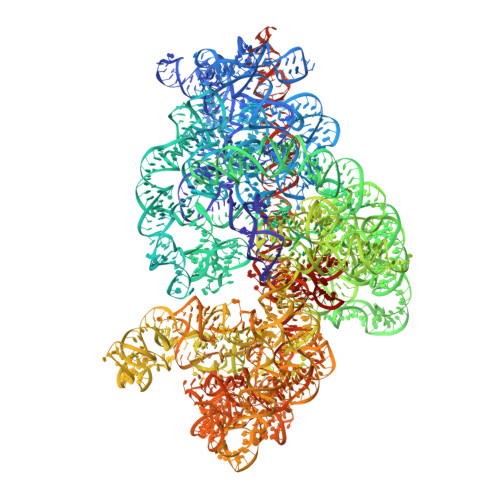
The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
Bieri, P., Leibundgut, M., Saurer, M., Boehringer, D., Ban, N.(2017) EMBO J 36: 475-486
- PubMed: 28007896
- DOI: https://doi.org/10.15252/embj.201695959
- Primary Citation of Related Structures:
5MMI, 5MMJ, 5MMM - PubMed Abstract:
Chloroplasts are cellular organelles of plants and algae that are responsible for energy conversion and carbon fixation by the photosynthetic reaction. As a consequence of their endosymbiotic origin, they still contain their own genome and the machinery for protein biosynthesis. Here, we present the atomic structure of the chloroplast 70S ribosome prepared from spinach leaves and resolved by cryo-EM at 3.4 Å resolution. The complete structure reveals the features of the 4.5S rRNA, which probably evolved by the fragmentation of the 23S rRNA, and all five plastid-specific ribosomal proteins. These proteins, required for proper assembly and function of the chloroplast translation machinery, bind and stabilize rRNA including regions that only exist in the chloroplast ribosome. Furthermore, the structure reveals plastid-specific extensions of ribosomal proteins that extensively remodel the mRNA entry and exit site on the small subunit as well as the polypeptide tunnel exit and the putative binding site of the signal recognition particle on the large subunit. The translation factor pY, involved in light- and temperature-dependent control of protein synthesis, is bound to the mRNA channel of the small subunit and interacts with 16S rRNA nucleotides at the A-site and P-site, where it protects the decoding centre and inhibits translation by preventing tRNA binding. The small subunit is locked by pY in a non-rotated state, in which the intersubunit bridges to the large subunit are stabilized.
Organizational Affiliation:
Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zurich, Zurich, Switzerland.