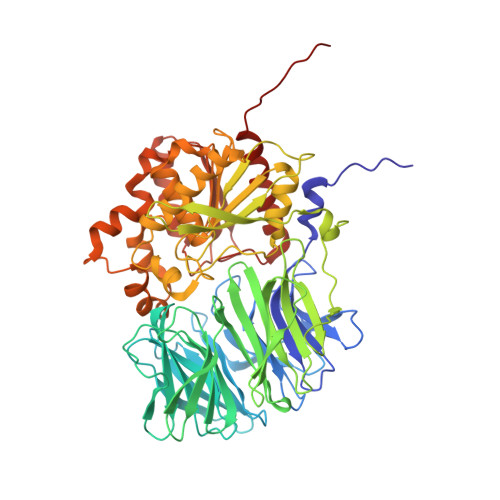
A bacterial acyl aminoacyl peptidase couples flexibility and stability as a result of cold adaptation.
Brocca, S., Ferrari, C., Barbiroli, A., Pesce, A., Lotti, M., Nardini, M.(2016) FEBS J 283: 4310-4324
- PubMed: 27739253
- DOI: https://doi.org/10.1111/febs.13925
- Primary Citation of Related Structures:
5L8S - PubMed Abstract:
Life in cold environments requires an overall increase in the flexibility of macromolecular and supramolecular structures to allow biological processes to take place at low temperature. Conformational flexibility supports high catalytic rates of enzymes in the cold but in several cases is also a cause of instability. The three-dimensional structure of the psychrophilic acyl aminoacyl peptidase from Sporosarcina psychrophila (SpAAP) reported in this paper highlights adaptive molecular changes resulting in a fine-tuned trade-off between flexibility and stability. In its functional form SpAAP is a dimer, and an increase in flexibility is achieved through loosening of intersubunit hydrophobic interactions. The release of subunits from the quaternary structure is hindered by an 'arm exchange' mechanism, in which a tiny structural element at the N terminus of one subunit inserts into the other subunit. Mutants lacking the 'arm' are monomeric, inactive and highly prone to aggregation. Another feature of SpAAP cold adaptation is the enlargement of the tunnel connecting the exterior of the protein with the active site. Such a wide channel might compensate for the reduced molecular motions occurring in the cold and allow easy and direct access of substrates to the catalytic site, rendering transient movements between domains unnecessary. Thus, cold-adapted SpAAP has developed a molecular strategy unique within this group of proteins: it is able to enhance the flexibility of each functional unit while still preserving sufficient stability. Structural data are available in the Protein Data Bank under the accession number 5L8S.
Organizational Affiliation:
Department of Biotechnology and Biosciences, University of Milano-Bicocca, Italy.