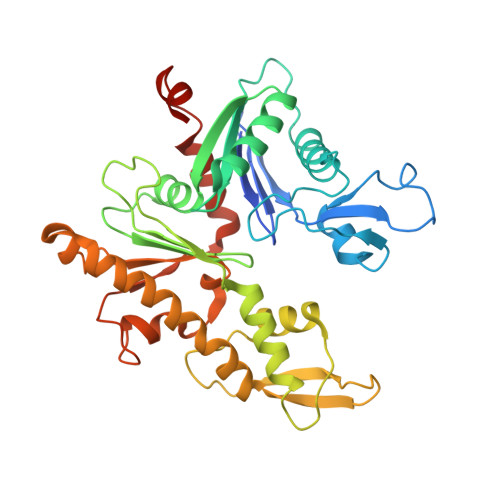
Structure of the magnetosome-associated actin-like MamK filament at subnanometer resolution.
Bergeron, J.R., Hutto, R., Ozyamak, E., Hom, N., Hansen, J., Draper, O., Byrne, M.E., Keyhani, S., Komeili, A., Kollman, J.M.(2017) Protein Sci 26: 93-102
- PubMed: 27391173
- DOI: https://doi.org/10.1002/pro.2979
- Primary Citation of Related Structures:
5JYG - PubMed Abstract:
Magnetotactic bacteria possess cellular compartments called magnetosomes that sense magnetic fields. Alignment of magnetosomes in the bacterial cell is necessary for their function, and this is achieved through anchoring of magnetosomes to filaments composed of the protein MamK. MamK is an actin homolog that polymerizes upon ATP binding. Here, we report the structure of the MamK filament at ∼6.5 Å, obtained by cryo-Electron Microscopy. This structure confirms our previously reported double-stranded, nonstaggered architecture, and reveals the molecular basis for filament formation. While MamK is closest in sequence to the bacterial actin MreB, the longitudinal contacts along each MamK strand most closely resemble those of eukaryotic actin. In contrast, the cross-strand interface, with a surprisingly limited set of contacts, is novel among actin homologs and gives rise to the nonstaggered architecture.
- Department of Biochemistry, University of Washington, Seattle, Washington.
Organizational Affiliation: