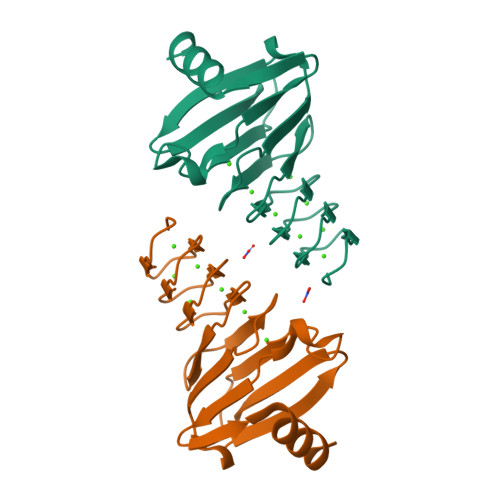
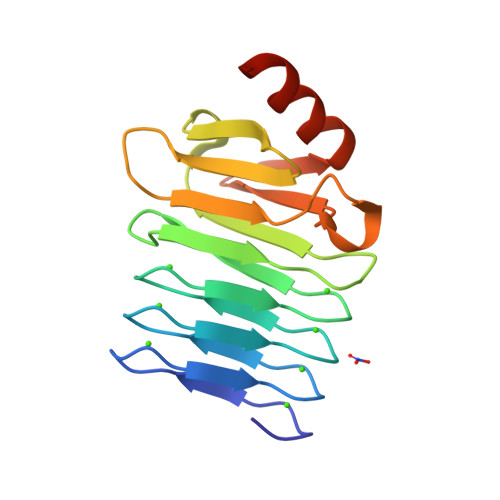
Calcium-Driven Folding of RTX Domain beta-Rolls Ratchets Translocation of RTX Proteins through Type I Secretion Ducts.
Bumba, L., Masin, J., Macek, P., Wald, T., Motlova, L., Bibova, I., Klimova, N., Bednarova, L., Veverka, V., Kachala, M., Svergun, D.I., Barinka, C., Sebo, P.(2016) Mol Cell 62: 47-62
- PubMed: 27058787
- DOI: https://doi.org/10.1016/j.molcel.2016.03.018
- Primary Citation of Related Structures:
5CVW, 5CXL - PubMed Abstract:
Calcium-binding RTX proteins are equipped with C-terminal secretion signals and translocate from the Ca(2+)-depleted cytosol of Gram-negative bacteria directly into the Ca(2+)-rich external milieu, passing through the "channel-tunnel" ducts of type I secretion systems (T1SSs). Using Bordetella pertussis adenylate cyclase toxin, we solved the structure of an essential C-terminal assembly that caps the RTX domains of RTX family leukotoxins. This is shown to scaffold directional Ca(2+)-dependent folding of the carboxy-proximal RTX repeat blocks into β-rolls. The resulting intramolecular Brownian ratchets then prevent backsliding of translocating RTX proteins in the T1SS conduits and thereby accelerate excretion of very large RTX leukotoxins from bacterial cells by a vectorial "push-ratchet" mechanism. Successive Ca(2+)-dependent and cosecretional acquisition of a functional RTX toxin structure in the course of T1SS-mediated translocation, through RTX domain folding from the C-terminal cap toward the N terminus, sets a paradigm that opens for design of virulence inhibitors of major pathogens.
Organizational Affiliation:
Institute of Microbiology, ASCR, v.v.i., Videnska 1083, 14220 Prague, Czech Republic.