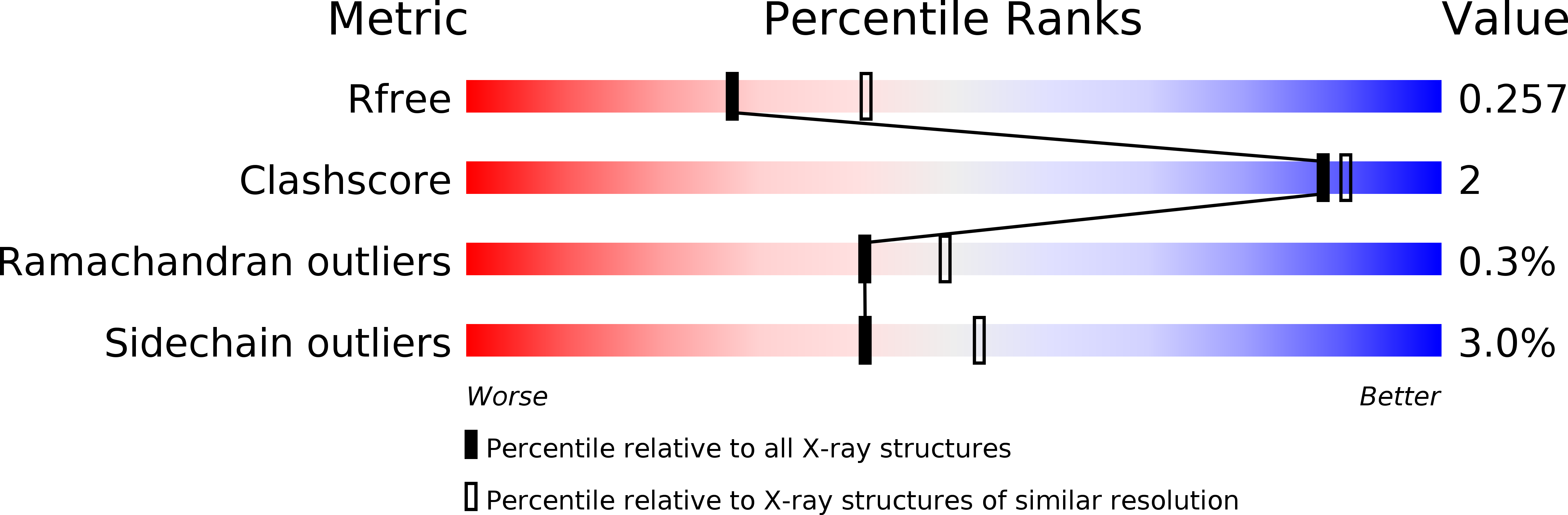
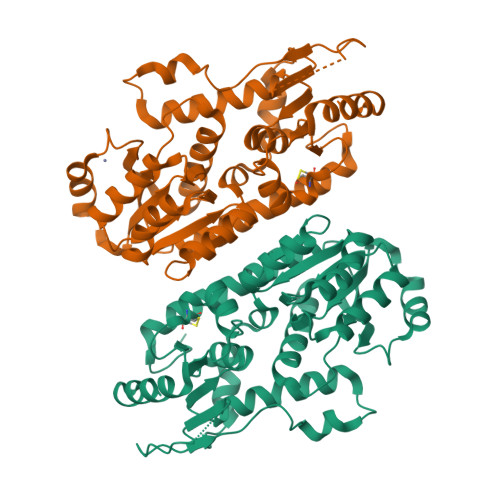
A rationally engineered yeast pyruvyltransferase Pvg1p introduces sialylation-like properties in neo-human-type complex oligosaccharide
Higuchi, Y., Yoshinaga, S., Yoritsune, K., Tateno, H., Hirabayashi, J., Nakakita, S., Kanekiyo, M., Kakuta, Y., Takegawa, K.(2016) Sci Rep 6: 26349-26349
- PubMed: 27194449
- DOI: https://doi.org/10.1038/srep26349
- Primary Citation of Related Structures:
5AX7 - PubMed Abstract:
Pyruvylation onto the terminus of oligosaccharide, widely seen from prokaryote to eukaryote, confers negative charges on the cell surface and seems to be functionally similar to sialylation, which is found at the end of human-type complex oligosaccharide. However, detailed molecular mechanisms underlying pyruvylation have not been clarified well. Here, we first determined the crystal structure of fission yeast pyruvyltransferase Pvg1p at a resolution of 2.46 Å. Subsequently, by combining molecular modeling with mutational analysis of active site residues, we obtained a Pvg1p mutant (Pvg1p(H168C)) that efficiently transferred pyruvyl moiety onto a human-type complex glycopeptide. The resultant pyruvylated human-type complex glycopeptide recognized similar lectins on lectin arrays as the α2,6-sialyl glycopeptides. This newly-generated pyruvylation of human-type complex oligosaccharides would provide a novel method for glyco-bioengineering.
Organizational Affiliation:
Department of Bioscience and Biotechnology, Faculty of Agriculture, Kyushu University, 6-10-1 Hakozaki, Fukuoka 812-8581, Japan.