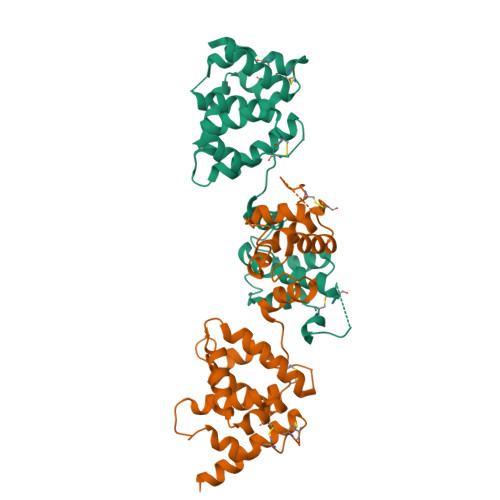
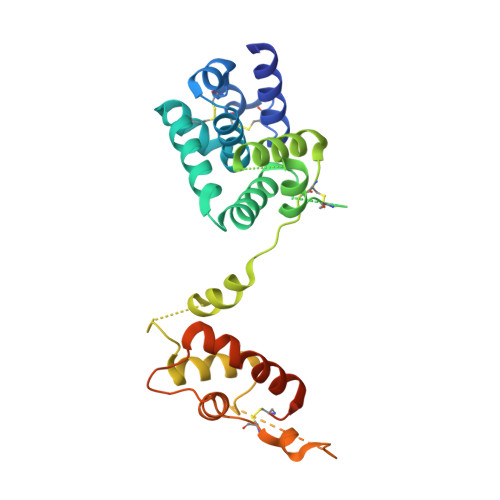
Structural insights of ZIP4 extracellular domain critical for optimal zinc transport.
Zhang, T., Sui, D., Hu, J.(2016) Nat Commun 7: 11979-11979
- PubMed: 27321477
- DOI: https://doi.org/10.1038/ncomms11979
- Primary Citation of Related Structures:
4X82 - PubMed Abstract:
The ZIP zinc transporter family is responsible for zinc uptake from the extracellular milieu or intracellular vesicles. The LIV-1 subfamily, containing nine out of the 14 human ZIP proteins, is featured with a large extracellular domain (ECD). The critical role of the ECD is manifested by disease-causing mutations on ZIP4, a representative LIV-1 protein. Here we report the first crystal structure of a mammalian ZIP4-ECD, which reveals two structurally independent subdomains and an unprecedented dimer centred at the signature PAL motif. Structure-guided mutagenesis, cell-based zinc uptake assays and mapping of the disease-causing mutations indicate that the two subdomains play pivotal but distinct roles and that the bridging region connecting them is particularly important for ZIP4 function. These findings lead to working hypotheses on how ZIP4-ECD exerts critical functions in zinc transport. The conserved dimeric architecture in ZIP4-ECD is also demonstrated to be a common structural feature among the LIV-1 proteins.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, Michigan 48824, USA.