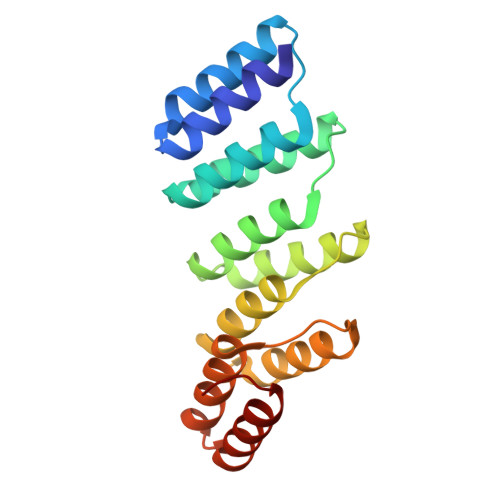
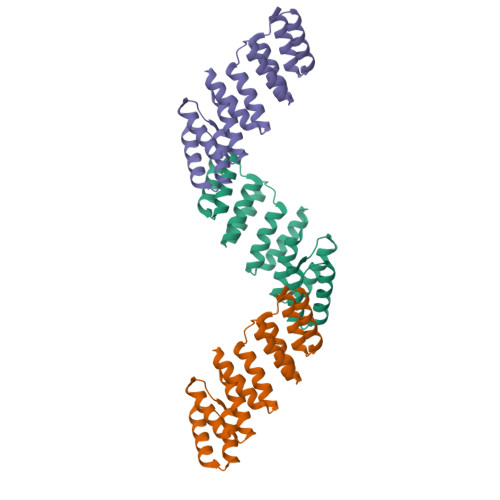
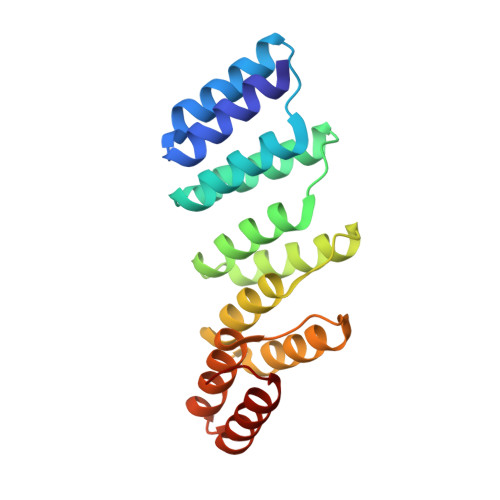
The design and structural characterization of a synthetic pentatricopeptide repeat protein.
Gully, B.S., Shah, K.R., Lee, M., Shearston, K., Smith, N.M., Sadowska, A., Blythe, A.J., Bernath-Levin, K., Stanley, W.A., Small, I.D., Bond, C.S.(2015) Acta Crystallogr D Biol Crystallogr 71: 196-208
- PubMed: 25664731
- DOI: https://doi.org/10.1107/S1399004714024869
- Primary Citation of Related Structures:
4OZS - PubMed Abstract:
Proteins of the pentatricopeptide repeat (PPR) superfamily are characterized by tandem arrays of a degenerate 35-amino-acid α-hairpin motif. PPR proteins are typically single-stranded RNA-binding proteins with essential roles in organelle biogenesis, RNA editing and mRNA maturation. A modular, predictable code for sequence-specific binding of RNA by PPR proteins has recently been revealed, which opens the door to the de novo design of bespoke proteins with specific RNA targets, with widespread biotechnological potential. Here, the design and production of a synthetic PPR protein based on a consensus sequence and the determination of its crystal structure to 2.2 Å resolution are described. The crystal structure displays helical disorder, resulting in electron density representing an infinite superhelical PPR protein. A structural comparison with related tetratricopeptide repeat (TPR) proteins, and with native PPR proteins, reveals key roles for conserved residues in directing the structure and function of PPR proteins. The designed proteins have high solubility and thermal stability, and can form long tracts of PPR repeats. Thus, consensus-sequence synthetic PPR proteins could provide a suitable backbone for the design of bespoke RNA-binding proteins with the potential for high specificity.
Organizational Affiliation:
School of Chemistry and Biochemistry, The University of Western Australia, Crawley, Western Australia, Australia.