Structure and Activity of the Flagellar Rotor Protein FliY: A MEMBER OF THE CheC PHOSPHATASE FAMILY.
Sircar, R., Greenswag, A.R., Bilwes, A.M., Gonzalez-Bonet, G., Crane, B.R.(2013) J Biological Chem 288: 13493-13502
- PubMed: 23532838
- DOI: https://doi.org/10.1074/jbc.M112.445171
- Primary Citation of Related Structures:
4HYN - PubMed Abstract:
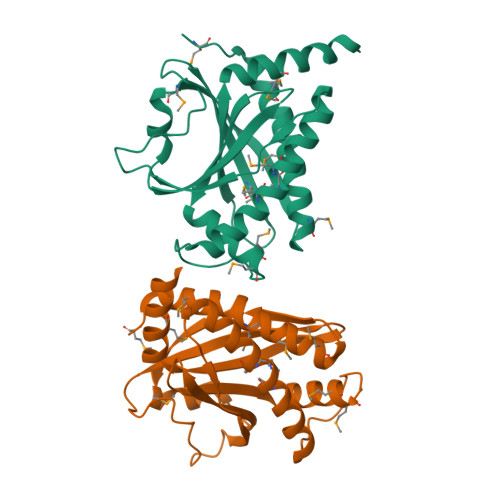
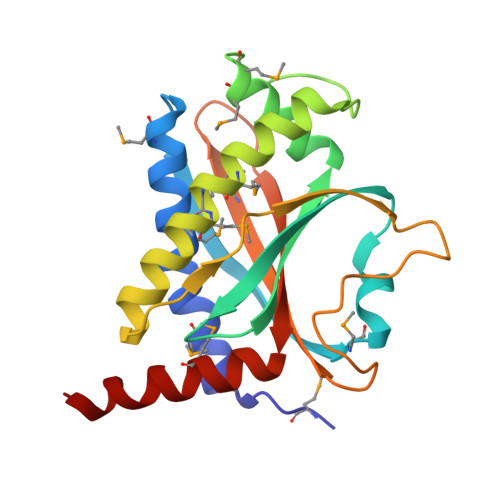
FliY is a flagellar rotor protein of the CheC phosphatase family. The FliY structure resembles that of the rotor protein FliM but contains two active centers for CheY dephosphorylation. FliY incorporates properties of the FliM/FliN rotor proteins and the CheC/CheX phosphatases to serve multiple functions in the flagellar switch. FliY distinguishes flagellar architecture and function in different types of bacteria. Rotating flagella propel bacteria toward favorable environments. Sense of rotation is determined by the intracellular response regulator CheY, which when phosphorylated (CheY-P) interacts directly with the flagellar motor. In many different types of bacteria, the CheC/CheX/FliY (CXY) family of phosphatases terminates the CheY-P signal. Unlike CheC and CheX, FliY is localized in the flagellar switch complex, which also contains the stator-coupling protein FliG and the target of CheY-P, FliM. The 2.5 Å resolution crystal structure of the FliY catalytic domain from Thermotoga maritima bears strong resemblance to the middle domain of FliM. Regions of FliM that mediate contacts within the rotor compose the phosphatase active sites in FliY. Despite the similarity between FliY and FliM, FliY does not bind FliG and thus is unlikely to be a substitute for FliM in the center of the switch complex. Solution studies indicate that FliY dimerizes through its C-terminal domains, which resemble the Escherichia coli switch complex component FliN. FliY differs topologically from the E. coli chemotaxis phosphatase CheZ but appears to utilize similar structural motifs for CheY dephosphorylation in close analogy to CheX. Recognition properties and phosphatase activities of site-directed mutants identify two pseudosymmetric active sites in FliY (Glu(35)/Asn(38) and Glu(132)/Asn(135)), with the second site (Glu(132)/Asn(135)) being more active. A putative N-terminal CheY binding domain conserved with FliM is not required for binding CheY-P or phosphatase activity.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14850, USA.