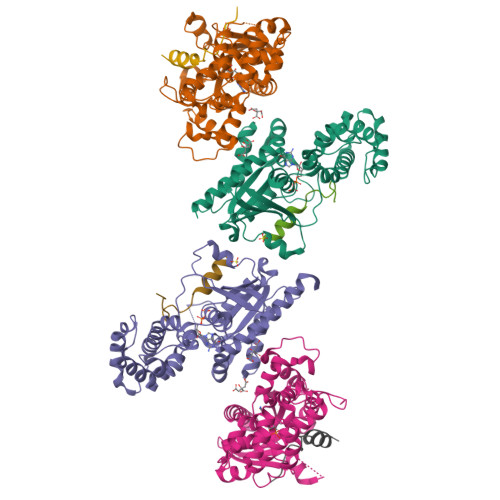
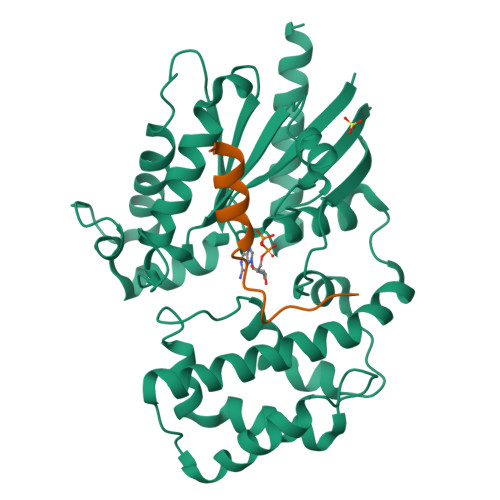
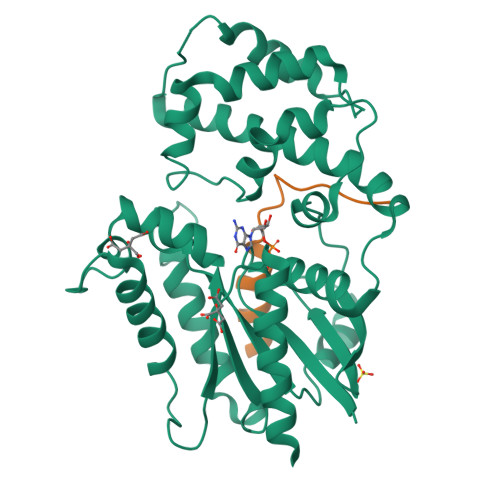
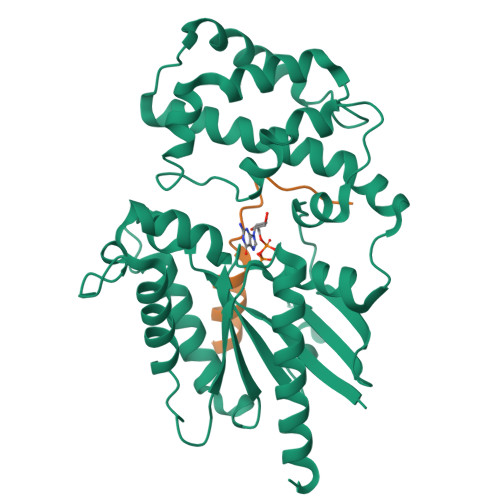
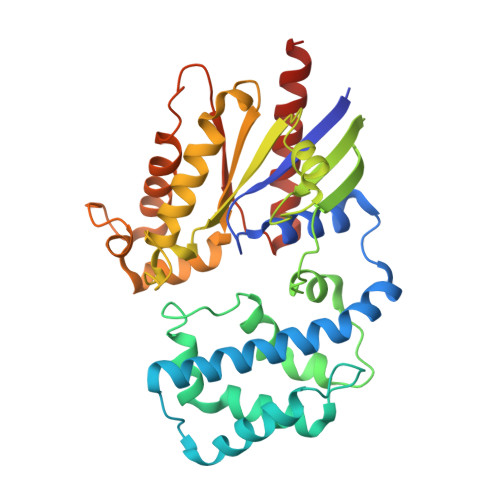
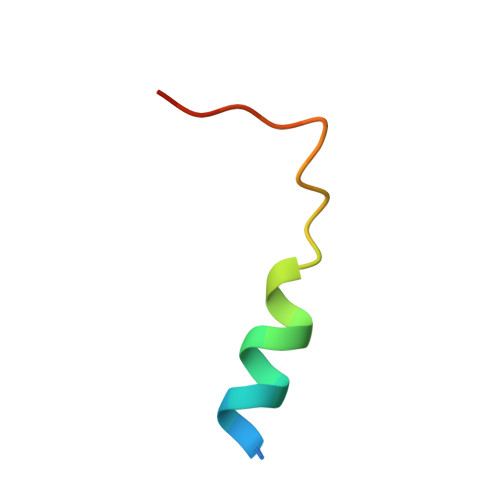
Crystal structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from G alpha i.
Jia, M., Li, J., Zhu, J., Wen, W., Zhang, M., Wang, W.(2012) J Biol Chem 287: 36766-36776
- PubMed: 22952234
- DOI: https://doi.org/10.1074/jbc.M112.391607
- Primary Citation of Related Structures:
4G5O, 4G5Q, 4G5R, 4G5S - PubMed Abstract:
GoLoco (GL) motif-containing proteins regulate G protein signaling by binding to Gα subunit and acting as guanine nucleotide dissociation inhibitors. GLs of LGN are also known to bind the GDP form of Gα(i/o) during asymmetric cell division. Here, we show that the C-terminal GL domain of LGN binds four molecules of Gα(i)·GDP. The crystal structures of Gα(i)·GDP in complex with LGN GL3 and GL4, respectively, reveal distinct GL/Gα(i) interaction features when compared with the only high resolution structure known with GL/Gα(i) interaction between RGS14 and Gα(i1.) Only a few residues C-terminal to the conserved GL sequence are required for LGN GLs to bind to Gα(i)·GDP. A highly conserved "double Arg finger" sequence (RΨ(D/E)(D/E)QR) is responsible for LGN GL to bind to GDP bound to Gα(i). Together with the sequence alignment, we suggest that the LGN GL/Gα(i) interaction represents a general binding mode between GL motifs and Gα(i). We also show that LGN GLs are potent guanine nucleotide dissociation inhibitors.
Organizational Affiliation:
Shanghai Key Laboratory of Molecular Catalysis and Innovative Materials, Department of Chemistry and Institute of Biomedical Sciences, Fudan University, Shanghai 200433, China.