Structural Analysis of Imp-L2 Function
Kulahin, N., Watson, C.J., Turkenburg, J.P., Kristensen, O., Norrman, M., Sajid, W., Schluckebier, G., Meyts, P.D., Brzozowski, M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NEURAL/ECTODERMAL DEVELOPMENT FACTOR IMP-L2 | 268 | Drosophila melanogaster | Mutation(s): 0 | 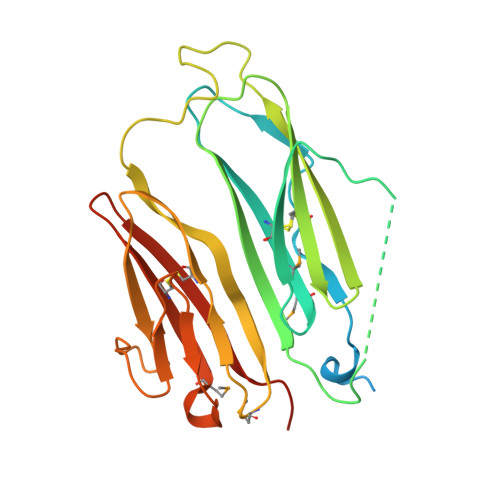 | |
UniProt | |||||
Find proteins for Q09024 (Drosophila melanogaster) Explore Q09024 Go to UniProtKB: Q09024 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q09024 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | C [auth A], D [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 49.856 | α = 90 |
| b = 99.732 | β = 91.99 |
| c = 56.28 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| MOSFLM | data reduction |
| xia2 | data scaling |
| Aimless | data scaling |
| HKL2Map | phasing |