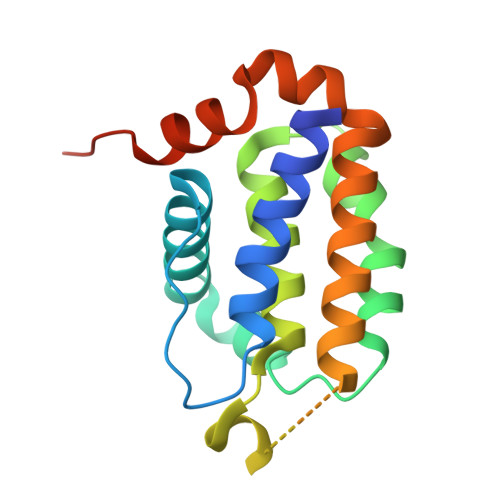
The Restricted Binding Repertoire of Bcl-B Leaves Bim as the Universal Bh3-Only Prosurvival Bcl-2 Protein Antagonist.
Rautureau, G.J.P., Yabal, M., Yang, H., Huang, D.C.S., Kvansakul, M., Hinds, M.G.(2012) Cell Death Dis 3: E443
- PubMed: 23235460
- DOI: https://doi.org/10.1038/cddis.2012.178
- Primary Citation of Related Structures:
4B4S - PubMed Abstract:
B-cell lymphoma-2 (Bcl-2) proteins mediate intrinsic-, or mitochondrial-, initiated apoptosis. We have investigated the structure and function of the least characterized Bcl-2 family member, Bcl-B, solving the crystal structure of a Bcl-B:Bim complex to 1.9 Å resolution. Bcl-B is distinguished from other Bcl-2 family members through an insertion of an unstructured loop between helices α5 and α6. Probing Bcl-B interactions with Bcl-2 homology (BH)3 motifs using a combination of biophysical- and cell-based assays revealed a unique BH3-only protein binding profile. Bcl-B has high-affinity interactions with Bim and Bik only. Our results not only delineate the mode of action of Bcl-B but also complete our understanding of the specific interactions between BH3-only proteins and their prosurvival Bcl-2 counterparts. Notably, we conclude that Bim is the universal prosurvival antagonist as no other BH3-only protein binds all six prosurvival proteins and that Mcl-1 and Bcl-x(L) form a distinct prosurvival dyad.
Organizational Affiliation:
Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3052, Australia.