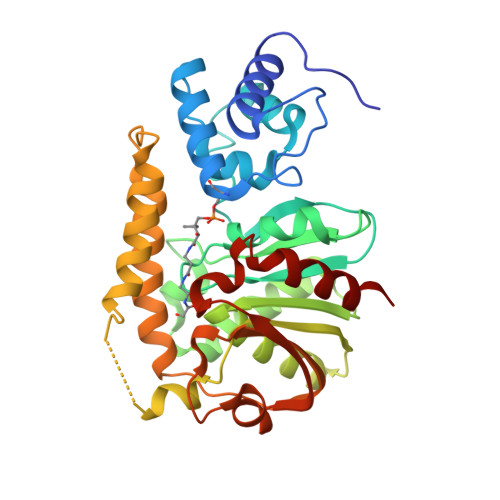
Structural basis for phosphopantetheinyl carrier domain interactions in the terminal module of nonribosomal peptide synthetases.
Liu, Y., Zheng, T., Bruner, S.D.(2011) Chem Biol 18: 1482-1488
- PubMed: 22118682
- DOI: https://doi.org/10.1016/j.chembiol.2011.09.018
- Primary Citation of Related Structures:
3TEJ - PubMed Abstract:
Phosphopantetheine-modified carrier domains play a central role in the template-directed, biosynthesis of several classes of primary and secondary metabolites. Fatty acids, polyketides, and nonribosomal peptides are constructed on multidomain enzyme assemblies using phosphopantetheinyl thioester-linked carrier domains to traffic and activate building blocks. The carrier domain is a dynamic component of the process, shuttling pathway intermediates to sequential enzyme active sites. Here, we report an approach to structurally fix carrier domain/enzyme constructs suitable for X-ray crystallographic analysis. The structure of a two-domain construct of Escherichia coli EntF was determined with a conjugated phosphopantetheinyl-based inhibitor. The didomain structure is locked in an active orientation relevant to the chemistry of nonribosomal peptide biosynthesis. This structure provides details into the interaction of phosphopantetheine arm with the carrier domain and the active site of the thioesterase domain.
- Department of Chemistry, Boston College, Chestnut Hill, MA 02167, USA.
Organizational Affiliation: