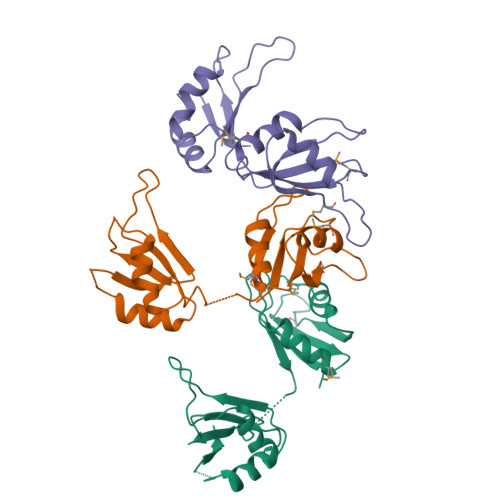
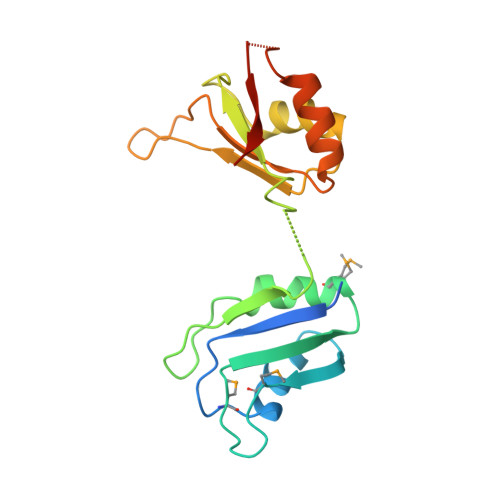
Absence of interdomain contacts in the crystal structure of the RNA recognition motifs of Sex-lethal.
Crowder, S.M., Kanaar, R., Rio, D.C., Alber, T.(1999) Proc Natl Acad Sci U S A 96: 4892-4897
- PubMed: 10220389
- DOI: https://doi.org/10.1073/pnas.96.9.4892
- Primary Citation of Related Structures:
3SXL - PubMed Abstract:
By binding specific RNA transcripts, the Sex-lethal protein (SXL) governs sexual differentiation and dosage compensation in Drosophila melanogaster. To investigate the basis for RNA binding specificity, we determined the crystal structure of the tandem RNA recognition motifs (RRMs) of SXL. Both RRMs adopt the canonical RRM fold, and the 10-residue, interdomain linker shows significant disorder. In contrast to the previously determined structure of the two-RRM fragment of heterogeneous nuclear ribonucleoprotein Al, SXL displays no interdomain contacts between RRMs. These results suggest that the SXL RRMs are flexibly tethered in solution, and RNA binding restricts the orientation of RRMs. Therefore, the observed specificity for single-stranded, U-rich sequences does not arise from a predefined, rigid architecture of the isolated SXL RRMs.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.