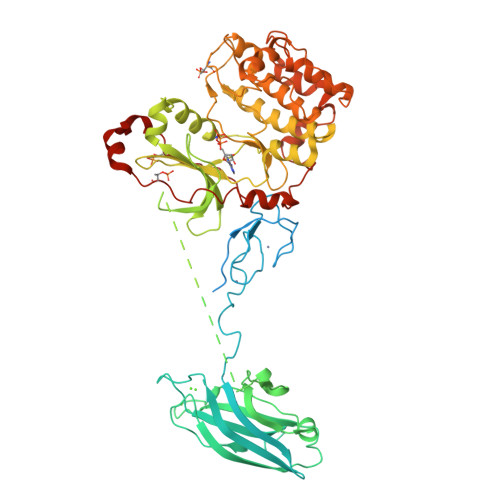
Crystal Structure and Allosteric Activation of Protein Kinase C beta II
Leonard, T.A., Rozycki, B., Saidi, L.F., Hummer, G., Hurley, J.H.(2011) Cell 144: 55-66
- PubMed: 21215369
- DOI: https://doi.org/10.1016/j.cell.2010.12.013
- Primary Citation of Related Structures:
3PFQ - PubMed Abstract:
Protein kinase C (PKC) isozymes are the paradigmatic effectors of lipid signaling. PKCs translocate to cell membranes and are allosterically activated upon binding of the lipid diacylglycerol to their C1A and C1B domains. The crystal structure of full-length protein kinase C βII was determined at 4.0 Å, revealing the conformation of an unexpected intermediate in the activation pathway. Here, the kinase active site is accessible to substrate, yet the conformation of the active site corresponds to a low-activity state because the ATP-binding side chain of Phe629 of the conserved NFD motif is displaced. The C1B domain clamps the NFD helix in a low-activity conformation, which is reversed upon membrane binding. A low-resolution solution structure of the closed conformation of PKCβII was derived from small-angle X-ray scattering. Together, these results show how PKCβII is allosterically regulated in two steps, with the second step defining a novel protein kinase regulatory mechanism.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institutes of Health, Bethesda, MD 20892, USA.