Structural insights into adeno-associated virus serotype 5.
Govindasamy, L., DiMattia, M.A., Gurda, B.L., Halder, S., McKenna, R., Chiorini, J.A., Muzyczka, N., Zolotukhin, S., Agbandje-McKenna, M.(2013) J Virol 87: 11187-11199
- PubMed: 23926356
- DOI: https://doi.org/10.1128/JVI.00867-13
- Primary Citation of Related Structures:
3NTT - PubMed Abstract:
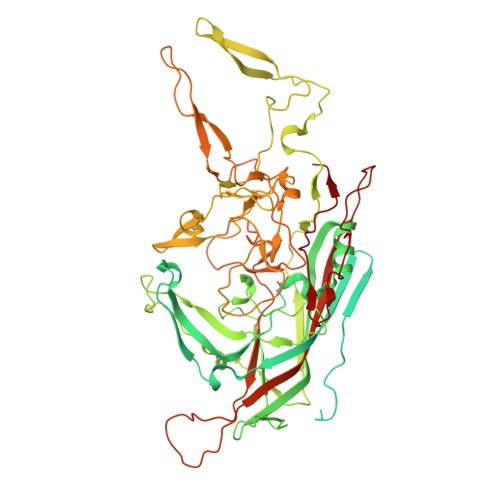
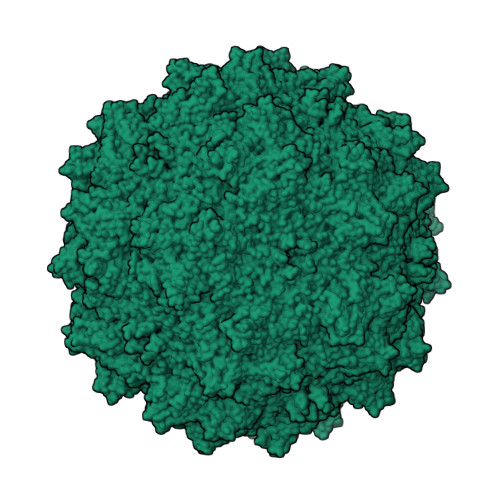
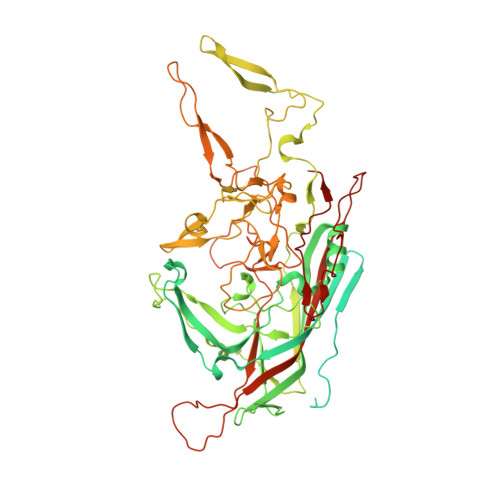
The adeno-associated viruses (AAVs) display differential cell binding, transduction, and antigenic characteristics specified by their capsid viral protein (VP) composition. Toward structure-function annotation, the crystal structure of AAV5, one of the most sequence diverse AAV serotypes, was determined to 3.45-Å resolution. The AAV5 VP and capsid conserve topological features previously described for other AAVs but uniquely differ in the surface-exposed HI loop between βH and βI of the core β-barrel motif and have pronounced conformational differences in two of the AAV surface variable regions (VRs), VR-IV and VR-VII. The HI loop is structurally conserved in other AAVs despite amino acid differences but is smaller in AAV5 due to an amino acid deletion. This HI loop is adjacent to VR-VII, which is largest in AAV5. The VR-IV, which forms the larger outermost finger-like loop contributing to the protrusions surrounding the icosahedral 3-fold axes of the AAVs, is shorter in AAV5, creating a smoother capsid surface topology. The HI loop plays a role in AAV capsid assembly and genome packaging, and VR-IV and VR-VII are associated with transduction and antigenic differences, respectively, between the AAVs. A comparison of interior capsid surface charge and volume of AAV5 to AAV2 and AAV4 showed a higher propensity of acidic residues but similar volumes, consistent with comparable DNA packaging capacities. This structure provided a three-dimensional (3D) template for functional annotation of the AAV5 capsid with respect to regions that confer assembly efficiency, dictate cellular transduction phenotypes, and control antigenicity.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Center for Structural Biology, McKnight Brain Institute, College of Medicine, University of Florida, Gainesville, Florida, USA.