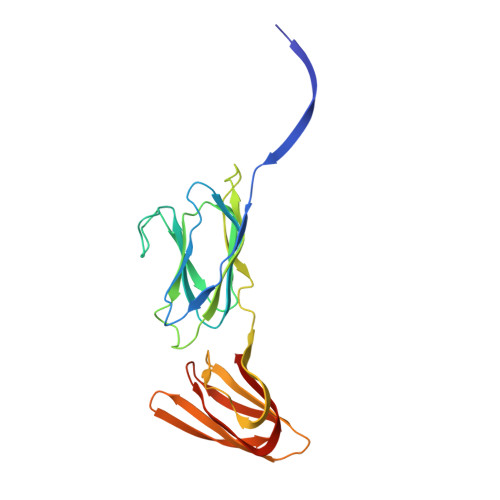
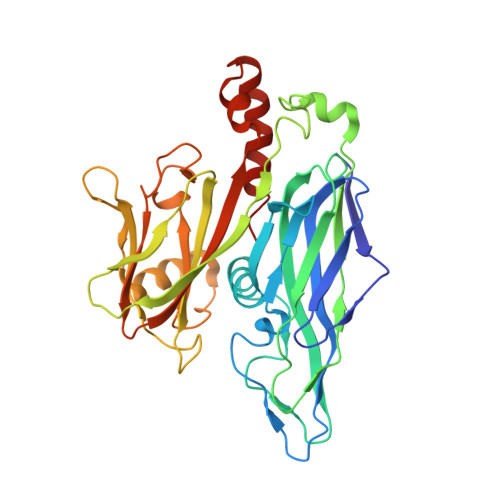
Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography.
Veesler, D., Ng, T.S., Sendamarai, A.K., Eilers, B.J., Lawrence, C.M., Lok, S.M., Young, M.J., Johnson, J.E., Fu, C.Y.(2013) Proc Natl Acad Sci U S A 110: 5504-5509
- PubMed: 23520050
- DOI: https://doi.org/10.1073/pnas.1300601110
- Primary Citation of Related Structures:
3J31, 4IL7 - PubMed Abstract:
Sulfolobus turreted icosahedral virus (STIV) was isolated in acidic hot springs where it infects the archeon Sulfolobus solfataricus. We determined the STIV structure using near-atomic resolution electron microscopy and X-ray crystallography allowing tracing of structural polypeptide chains and visualization of transmembrane proteins embedded in the viral membrane. We propose that the vertex complexes orchestrate virion assembly by coordinating interactions of the membrane and various protein components involved. STIV shares the same coat subunit and penton base protein folds as some eukaryotic and bacterial viruses, suggesting that they derive from a common ancestor predating the divergence of the three kingdoms of life. One architectural motif (β-jelly roll fold) forms virtually the entire capsid (distributed in three different gene products), indicating that a single ancestral protein module may have been at the origin of its evolution.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, CA, USA.
Organizational Affiliation: