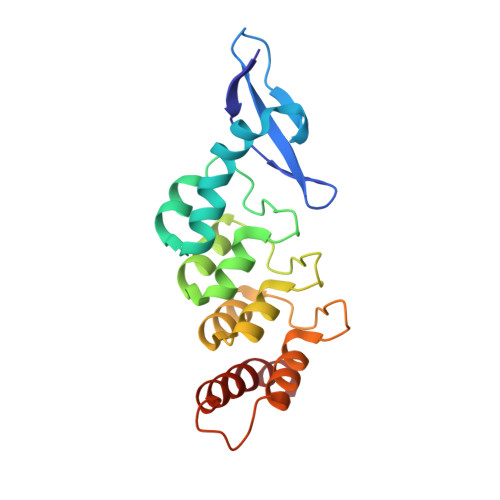
Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Stengel, K.F., Holdermann, I., Cain, P., Robinson, C., Wild, K., Sinning, I.(2008) Science 321: 253-256
- PubMed: 18621669
- DOI: https://doi.org/10.1126/science.1158640
- Primary Citation of Related Structures:
3DEO, 3DEP - PubMed Abstract:
Secretory and membrane proteins carry amino-terminal signal sequences that, in cotranslational targeting, are recognized by the signal recognition particle protein SRP54 without sequence specificity. The most abundant membrane proteins on Earth are the light-harvesting chlorophyll a/b binding proteins (LHCPs). They are synthesized in the cytoplasm, imported into the chloroplast, and posttranslationally targeted to the thylakoid membrane by cpSRP, a heterodimer formed by cpSRP54 and cpSRP43. We present the 1.5 angstrom crystal structure of cpSRP43 characterized by a unique arrangement of chromodomains and ankyrin repeats. The overall shape and charge distribution of cpSRP43 resembles the SRP RNA, which is absent in chloroplasts. The complex with the internal signal sequence of LHCPs reveals that cpSRP43 specifically recognizes a DPLG peptide motif. We describe how cpSPR43 adapts the universally conserved SRP system to posttranslational targeting and insertion of the LHCP family of membrane proteins.
Organizational Affiliation:
Biochemie-Zentrum der Universität Heidelberg, INF328, D-69120 Heidelberg, Germany.