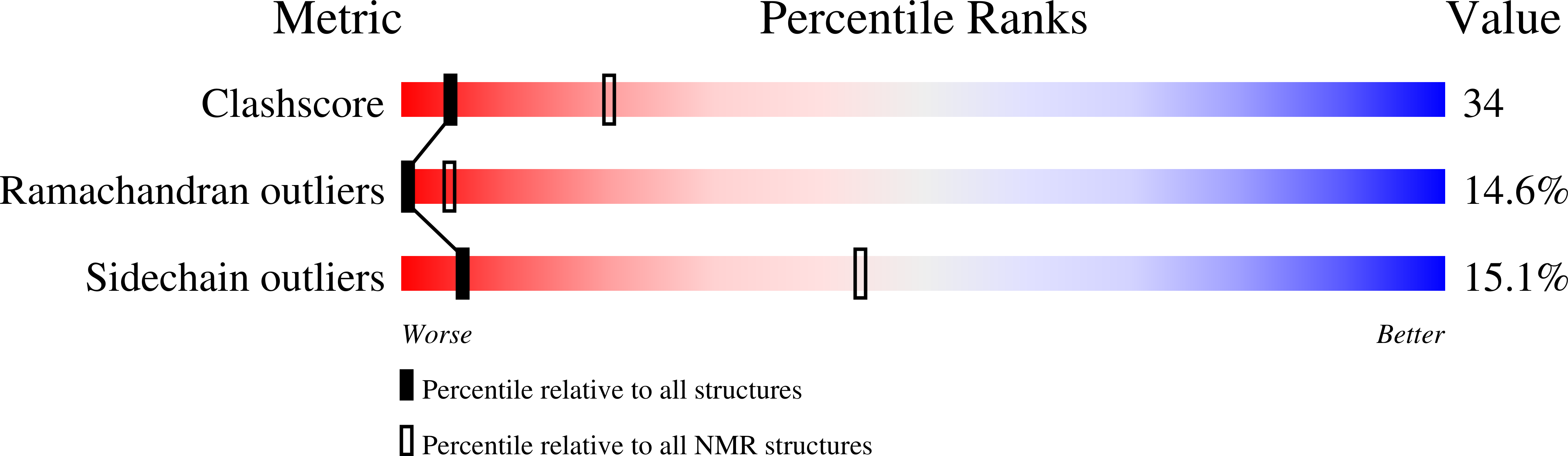The NMR solution structure of human epidermal growth factor (hEGF) at physiological pH and its interactions with suramin
Huang, H.W., Mohan, S.K., Yu, C.(2010) Biochem Biophys Res Commun 402: 705-710
- PubMed: 21029725
- DOI: https://doi.org/10.1016/j.bbrc.2010.10.089
- Primary Citation of Related Structures:
2KV4 - PubMed Abstract:
Human epidermal growth factor (hEGF) induces the proliferation, differentiation and survival of various cell types including tumor-derived cells. Generally, hEGF performs its biological function by binding to a specific receptor (hEGFR) on the cell surface, thereby inducing signal transduction. Suramin, a polysulfonated naphthylurea that acts as a growth factor blocker, exhibits antiproliferative activity against non-small cell lung cancer (NSCLC) cells that overexpress EGFR on the cell surface. We determined the solution structure of hEGF under physiological conditions and investigated the interaction of suramin with hEGF using isothermal titration calorimetry and NMR spectroscopy techniques. The solution structure of hEGF presented in this paper is different from the bound form of hEGF present in the crystal structure of the 2:2 EGF-EGFR complex because its C-tail contains a hydrophobic core. This conformational difference supports the hypothesis that hEGF undergoes a conformational change when it binds to hEGFR and subsequently induces signal transduction. Based on the docking structure of the hEGF-suramin complex, we demonstrated how suramin blocks hEGF by binding to its receptor binding site (the C-terminal region around Arg45) and inhibits the crucial conformational change.
Organizational Affiliation:
Department of Chemistry, National Tsing Hua University, Taiwan, ROC. d9623541@oz.nthu.edu.tw