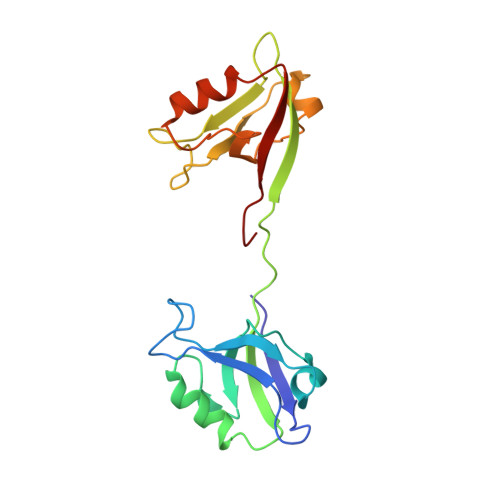
Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
Wang, W.N., Weng, J.W., Zhang, X., Liu, M.L., Zhang, M.J.(2009) J Am Chem Soc 131: 787-796
- PubMed: 19072119
- DOI: https://doi.org/10.1021/ja8076022
- Primary Citation of Related Structures:
2KA9 - PubMed Abstract:
The two N-terminal PDZ domains of postsynaptic density protein-95 (PDS-95 PDZ1 and PDZ2) are closely connected in tandem by a conserved peptide linker of five amino acids. The interdomain orientation between PDZ1 and PDZ2 of the ligand-free PDZ12 tandem is restrained, and this conformational arrangement facilitates the synergistic binding of PDZ12 to multimeric targets. (1) The interdomain orientation of the target-bound state of PDZ12 is not known. Here, we have solved the structure of PDZ12 in complex with its binding domain from cypin. Both chemical shift data and residual dipolar coupling measurements showed that the restrained interdomain orientation disappeared upon cypin peptide binding. NMR-based relaxation experiments revealed slow interdomain motions in the PDZ12/cypin peptide complex. Molecular dynamics simulations also showed that the PDZ12/cypin complex has larger conformational flexibility than the ligand-free PDZ12. This dramatic change of protein dynamics provides extra conformational entropy upon ligand binding, thus enhancing the ligand binding affinity of the PDZ12 tandem. Modulation of ligand binding affinity through concerted interdomain structural and dynamic rearrangements may represent a general property of multidomain scaffold proteins.
Organizational Affiliation:
Department of Chemistry, Fudan University, Shanghai, 200433, China. wnwang@fudan.edu.cn