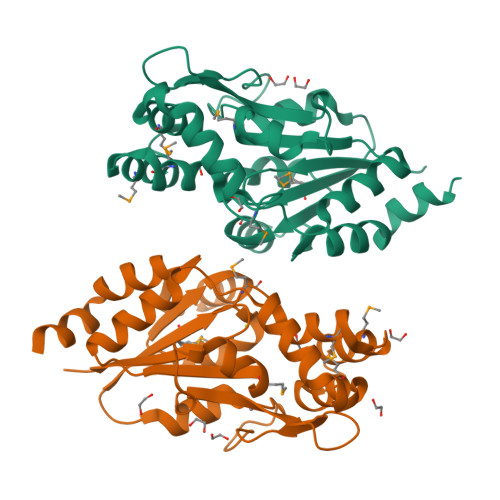
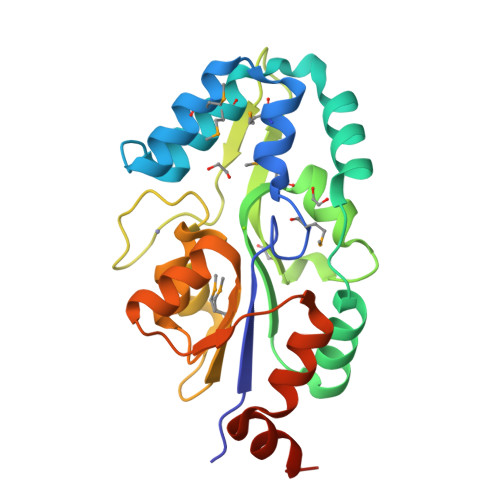
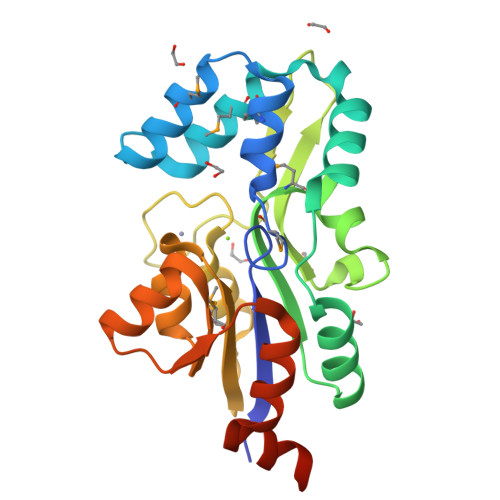
Crystal structure of MtnX phosphatase from Bacillus subtilis at 2.0 A resolution provides a structural basis for bipartite phosphomonoester hydrolysis of 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate.
Xu, Q., Saikatendu, K.S., Krishna, S.S., McMullan, D., Abdubek, P., Agarwalla, S., Ambing, E., Astakhova, T., Axelrod, H.L., Carlton, D., Chiu, H.J., Clayton, T., DiDonato, M., Duan, L., Elsliger, M.A., Feuerhelm, J., Grzechnik, S.K., Hale, J., Hampton, E., Han, G.W., Haugen, J., Jaroszewski, L., Jin, K.K., Klock, H.E., Knuth, M.W., Koesema, E., Miller, M.D., Morse, A.T., Nigoghossian, E., Okach, L., Oommachen, S., Paulsen, J., Reyes, R., Rife, C.L., Schwarzenbacher, R., van den Bedem, H., White, A., Wolf, G., Hodgson, K.O., Wooley, J., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A.(2007) Proteins 69: 433-439
- PubMed: 17654724
- DOI: https://doi.org/10.1002/prot.21602
- Primary Citation of Related Structures:
2FEA
Organizational Affiliation:
Stanford Synchrotron Radiation Laboratory, Stanford University, Menlo Park, California, USA.