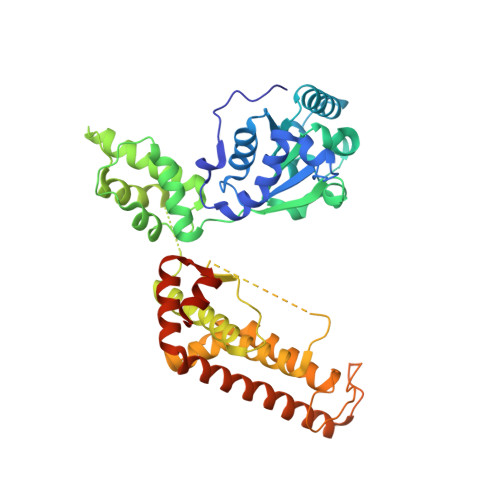
The Molecular Architecture of the Metalloprotease Ftsh.
Bieniossek, C., Schalch, T., Bumann, M., Meister, M., Meier, R., Baumann, U.(2006) Proc Natl Acad Sci U S A 103: 3066
- PubMed: 16484367
- DOI: https://doi.org/10.1073/pnas.0600031103
- Primary Citation of Related Structures:
2CE7, 2CEA - PubMed Abstract:
The ATP-dependent integral membrane protease FtsH is universally conserved in bacteria. Orthologs exist in chloroplasts and mitochondria, where in humans the loss of a close FtsH-homolog causes a form of spastic paraplegia. FtsH plays a crucial role in quality control by degrading unneeded or damaged membrane proteins, but it also targets soluble signaling factors like sigma(32) and lambda-CII. We report here the crystal structure of a soluble FtsH construct that is functional in caseinolytic and ATPase assays. The molecular architecture of this hexameric molecule consists of two rings where the protease domains possess an all-helical fold and form a flat hexagon that is covered by a toroid built by the AAA domains. The active site of the protease classifies FtsH as an Asp-zincin, contrary to a previous report. The different symmetries of protease and AAA rings suggest a possible translocation mechanism of the target polypeptide chain into the interior of the molecule where the proteolytic sites are located.
Organizational Affiliation:
Departement für Chemie und Biochemie, Universität Bern, Switzerland.