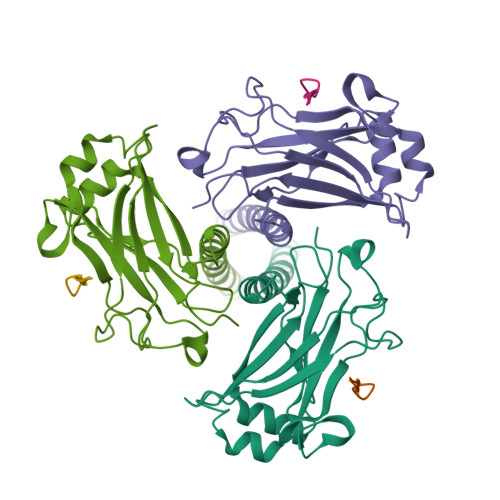
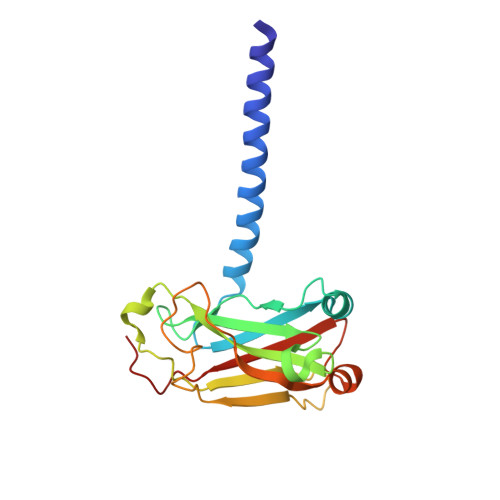
LMP1 protein from the Epstein Barr virus is a structural CD40 decoy in B lymphocytes fro binding to TRAF3
Wu, S.D., Xie, P., Welsh, K., Li, C., Ni, C.-Z., Zhu, X., Reed, J.C., Satterthwait, A.C., Bishop, G.A., Ely, K.R.(2005) J Biological Chem 39: 33620-33626
- PubMed: 16009714
- DOI: https://doi.org/10.1074/jbc.M502511200
- Primary Citation of Related Structures:
1ZMS - PubMed Abstract:
Epstein-Barr virus is a human herpesvirus that causes infectious mononucleosis and lymphoproliferative malignancies. LMP1 (latent membrane protein-1), which is encoded by this virus and which is essential for transformation of B lymphocytes, acts as a constitutively active mimic of the tumor necrosis factor receptor (TNFR) CD40. LMP1 is an integral membrane protein containing six transmembrane segments and a cytoplasmic domain at the C terminus that binds to intracellular TNFR-associated factors (TRAFs). TRAFs are intracellular co-inducers of downstream signaling from CD40 and other TNFRs, and TRAF3 is required for activation of B lymphocytes by LMP1. Cytoplasmic C-terminal activation region 1 of LMP1 bears a motif (PQQAT) that conforms to the TRAF recognition motif PVQET in CD40. In this study, we report the crystal structure of this portion of LMP1 C-terminal activation region-1 (204PQQATDD210) bound in complex with TRAF3. The PQQAT motif is bound in the same binding crevice on TRAF3 where CD40 is bound, providing a molecular mechanism for LMP1 to act as a CD40 decoy for TRAF3. The LMP1 motif is presented in the TRAF3 crevice as a close structural mimic of the PVQET motif in CD40, and the intermolecular contacts are similar. However, the viral protein makes a unique contact: a hydrogen bond network formed between Asp210 in LMP1 and Tyr395 and Arg393 in TRAF3. This intermolecular contact is not made in the CD40-TRAF3 complex. The additional hydrogen bonds may stabilize the complex and strengthen the binding to permit LMP1 to compete with CD40 for binding to the TRAF3 crevice, influencing downstream signaling to B lymphocytes and contributing to dysregulated signaling by LMP1.
Organizational Affiliation:
Cancer Center, The Burnham Institute, La Jolla, California 92037, USA.