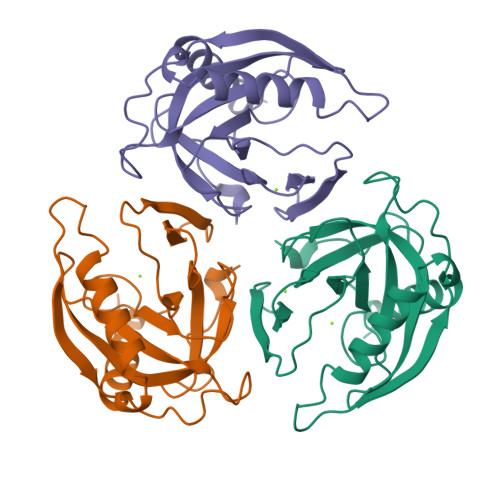
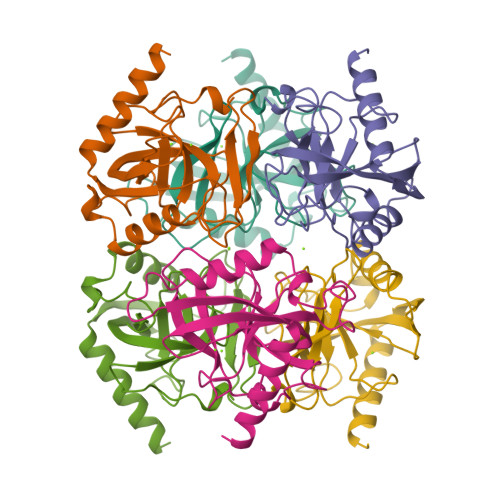
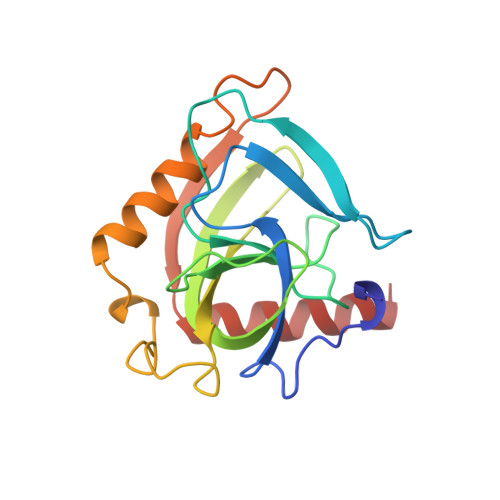
Crystal structure of holo inorganic pyrophosphatase from Escherichia coli at 1.9 A resolution. Mechanism of hydrolysis.
Harutyunyan, E.H., Oganessyan, V.Y., Oganessyan, N.N., Avaeva, S.M., Nazarova, T.I., Vorobyeva, N.N., Kurilova, S.A., Huber, R., Mather, T.(1997) Biochemistry 36: 7754-7760
- PubMed: 9201917
- DOI: https://doi.org/10.1021/bi962637u
- Primary Citation of Related Structures:
1OBW - PubMed Abstract:
Crystalline holo inorganic pyrophosphatase from Escherichia coli was grown in the presence of 250 mM MgCl2. The crystal structure has been solved by Patterson search techniques and refined to an R-factor of 17.6% at 1.9 A resolution. The upper estimate of the root-mean-square error in atomic positions is 0.26 A. These crystals belong to space group P3(2)21 with unit cell dimensions a = b = 110.27 A and c = 78.17 A. The asymmetric unit contains a trimer of subunits, i.e., half of the hexameric molecule. In the central cavity of the enzyme molecule, three Mg2+ ions, each shared by two subunits of the hexamer, are found. In the active sites of two crystallographically independent subunits, two Mg2+ ions are bound. The second active site Mg2+ ion is missing in the third subunit. A mechanism of catalysis is proposed whereby a water molecule activated by a Mg2+ ion and Tyr 55 play essential roles.
Organizational Affiliation:
Shubnikov Institute of Crystallography, Russian Academy of Sciences, Moscow, Russian Federation. emil@protein.crystal.msk.ru