Structural basis for recognition of antihistamine drug by human histamine receptor.
Peng, X., Yang, L., Liu, Z., Lou, S., Mei, S., Li, M., Chen, Z., Zhang, H.(2022) Nat Commun 13: 6105-6105
- PubMed: 36243875
- DOI: https://doi.org/10.1038/s41467-022-33880-y
- Primary Citation of Related Structures:
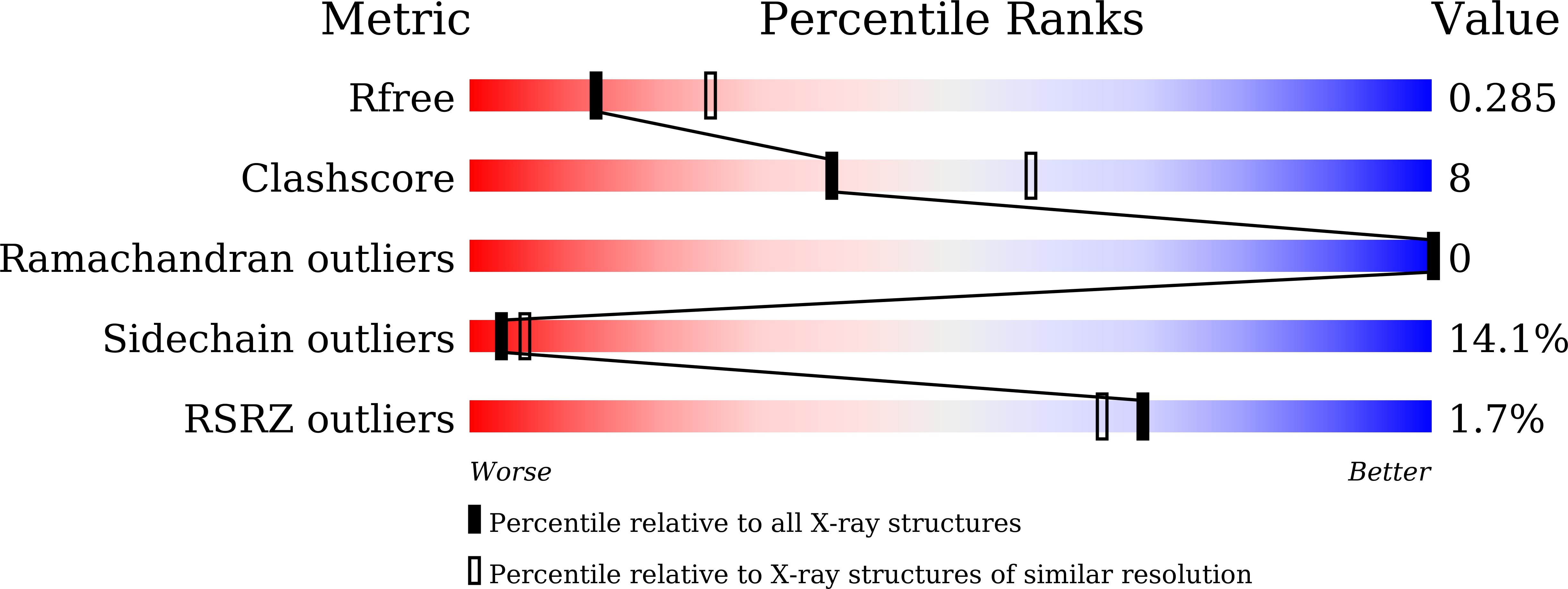
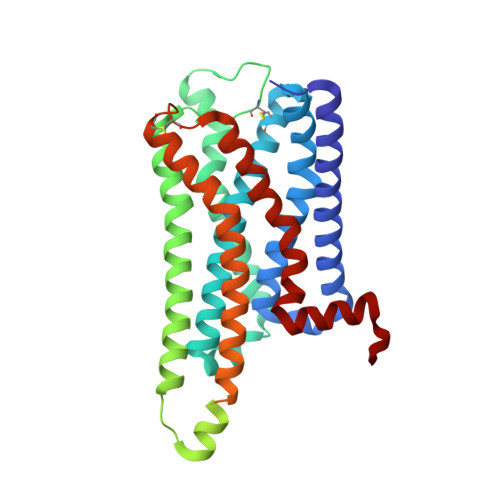
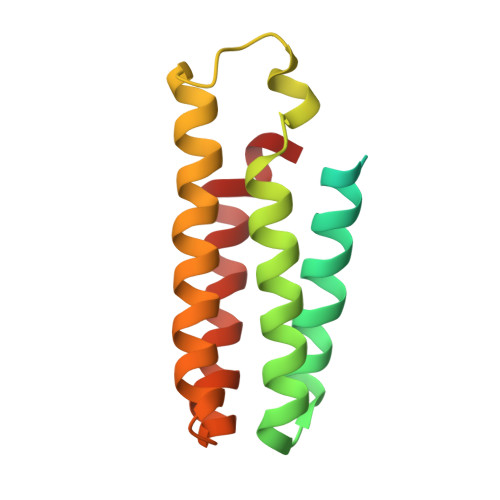
7F61 - PubMed Abstract:
The histamine receptors belong to the G protein-coupled receptor (GPCR) superfamily, and play important roles in the regulation of histamine and other neurotransmitters in the central nervous system, as potential targets for the treatment of neurologic and psychiatric disorders. Here we report the crystal structure of human histamine receptor H 3 R bound to an antagonist PF-03654746 at 2.6 Å resolution. Combined with the computational and functional assays, our structure reveals binding modes of the antagonist and allosteric cholesterol. Molecular dynamic simulations and molecular docking of different antihistamines further elucidate the conserved ligand-binding modes. These findings are therefore expected to facilitate the structure-based design of novel antihistamines.
Organizational Affiliation:
Hangzhou Institute of Innovative Medicine, Institute of Pharmacology and Toxicology, Zhejiang Province Key Laboratory of Anti-Cancer Drug Research, College of Pharmaceutical Sciences, Zhejiang University, 310058, Hangzhou, Zhejiang, China.