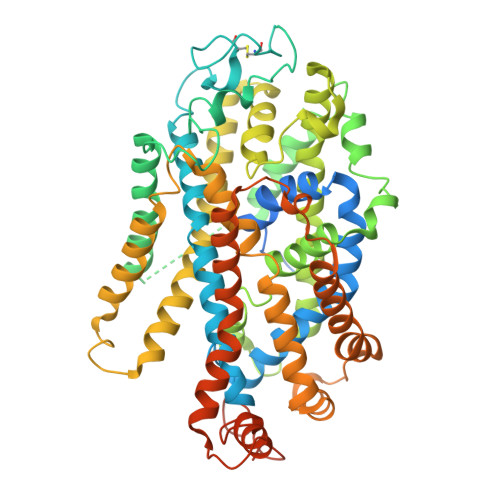
Modulation of the human GlyT1 by clinical drugs and cholesterol.
Li, N., Wei, Y., Li, R., Meng, Y., Zhao, J., Bai, Q., Wang, G., Zhao, Y.(2025) Nat Commun 16: 2412-2412
- PubMed: 40069141
- DOI: https://doi.org/10.1038/s41467-025-57613-z
- Primary Citation of Related Structures:
9J8B, 9J8C, 9J8D - PubMed Abstract:
Glycine transporter 1 (GlyT1) is a key player in shaping extracellular glutamatergic signaling processes and holds promise for treating cognitive impairments associated with schizophrenia by inhibiting its activity and thus enhancing the function of NMDA receptors. Despite its significant role in physiological and pharmacology, its modulation mechanism by clinical drugs and internal lipids remains elusive. Here, we determine cryo-EM structures of GlyT1 in its apo state and in complex with clinical trial drugs iclepertin and sarcosine. The GlyT1 in its apo state is determined in three distinct conformations, exhibiting a conformational equilibrium of the transport cycle. The complex structures with inhibitor iclepertin and sarcosine elucidate their unique binding poses with GlyT1. Three binding sites of cholesterol are determined in GlyT1, two of which are conformation-dependent. Transport kinetics studies reveal that a delicate binding equilibrium for cholesterol is crucial for the conformational transition of GlyT1. This study significantly enhances our understanding of the physiological and pharmacological aspects of GlyT1.
Organizational Affiliation:
Heart Center and Beijing Key Laboratory of Hypertension, Beijing Chaoyang Hospital, Capital Medical University, Beijing, China.