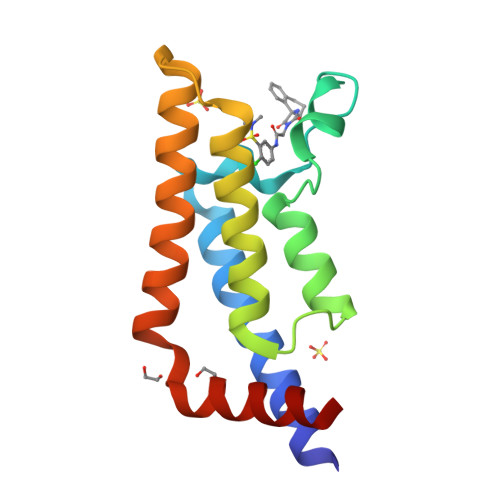
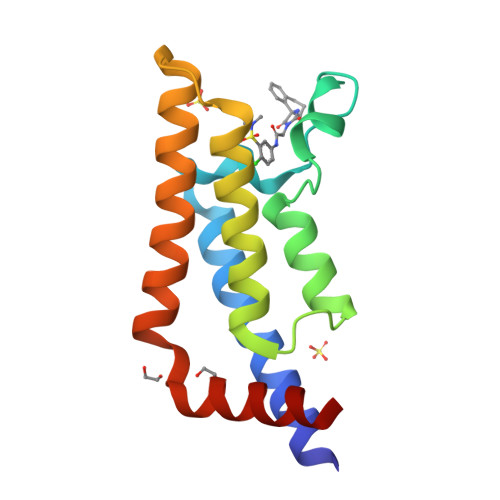
A Qualified Success: Discovery of a New Series of ATAD2 Bromodomain Inhibitors with a Novel Binding Mode Using High-Throughput Screening and Hit Qualification.
Bamborough, P., Chung, C.W., Demont, E.H., Bridges, A.M., Craggs, P.D., Dixon, D.P., Francis, P., Furze, R.C., Grandi, P., Jones, E.J., Karamshi, B., Locke, K., Lucas, S.C.C., Michon, A.M., Mitchell, D.J., Pogany, P., Prinjha, R.K., Rau, C., Roa, A.M., Roberts, A.D., Sheppard, R.J., Watson, R.J.(2019) J Med Chem 62: 7506-7525
- PubMed: 31398032
- DOI: https://doi.org/10.1021/acs.jmedchem.9b00673
- Primary Citation of Related Structures:
6S55, 6S56, 6S57 - PubMed Abstract:
The bromodomain of ATAD2 has proved to be one of the least-tractable proteins within this target class. Here, we describe the discovery of a new class of inhibitors by high-throughput screening and show how the difficulties encountered in establishing a screening triage capable of finding progressible hits were overcome by data-driven optimization. Despite the prevalence of nonspecific hits and an exceptionally low progressible hit rate (0.001%), our optimized hit qualification strategy employing orthogonal biophysical methods enabled us to identify a single active series. The compounds have a novel ATAD2 binding mode with noncanonical features including the displacement of all conserved water molecules within the active site and a halogen-bonding interaction. In addition to reporting this new series and preliminary structure-activity relationship, we demonstrate the value of diversity screening to complement the knowledge-based approach used in our previous ATAD2 work. We also exemplify tactics that can increase the chance of success when seeking new chemical starting points for novel and less-tractable targets.
Organizational Affiliation:
Cellzome , Meyerhofstrasse 1 , Heidelberg 69117 , Germany.