Crystal structure of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase, an NAD(+)-dependent dismutase from Mesorhizobium loti
Mugo, A.N., Kobayashi, J., Mikami, B., Yoshikane, Y., Yagi, T., Ohnishi, K.(2015) Biochem Biophys Res Commun 456: 35-40
- PubMed: 25446130
- DOI: https://doi.org/10.1016/j.bbrc.2014.11.028
- Primary Citation of Related Structures:
4OM8 - PubMed Abstract:
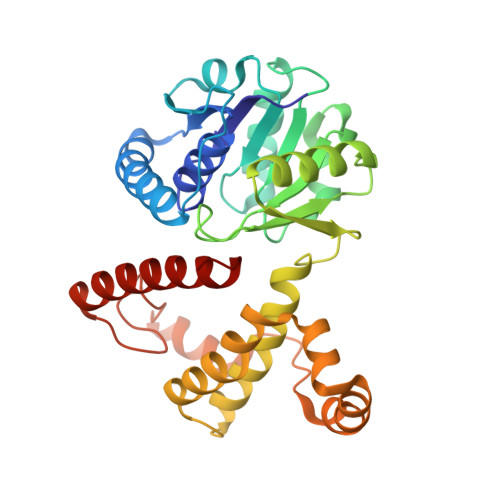
5-Formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase (FHMPCDH) from Mesorhizobium loti is the fifth enzyme in degradation pathway I for pyridoxine. The enzyme catalyzes a dismutation reaction: the oxidation of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid (FHMPC) to 3-hydroxy-2-methylpyridine 4,5-dicarboxylic acid with NAD(+) and reduction of FHMPC to 4-pyridoxic acid with NADH. FHMPCDH belongs to the l-3-hydroxyacyl-CoA dehydrogenase (HAD) family. The crystal structure was determined by molecular replacement and refined to a resolution of 1.55Å (R-factor of 16.4%, Rfree=19.4%). There were two monomers in the asymmetric unit. The overall structure of the monomer consisted of N- and C-terminal domains connected by a short linker loop. The monomer was similar to members of the HAD family (RMSD=1.9Å). The active site was located between the domains and highly conserved to that of human heart l-3-hydroxyacyl-CoA dehydrogenase (HhHAD). His-Glu catalytic dyad, a serine and two asparagine residues of HhHAD were conserved. Ser116, His137 and Glu149 in FHMPCDH are connected by a hydrogen bonding network forming a catalytic triad. The functions of the active site residues in the reaction mechanism are discussed.
Organizational Affiliation:
Research Institute of Molecular Genetics, Kochi University, Nankoku, Kochi 783-8502, Japan.