Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Zou, Y., Li, L., Chen, W.Y., Chen, T.T., Ma, L., Wang, X., Xiong, B., Xu, Y.C., Shen, J.(2013) Molecules 18: 5706-5722
- PubMed: 23681056
- DOI: https://doi.org/10.3390/molecules18055706
- Primary Citation of Related Structures:
4IVS, 4IVT - PubMed Abstract:
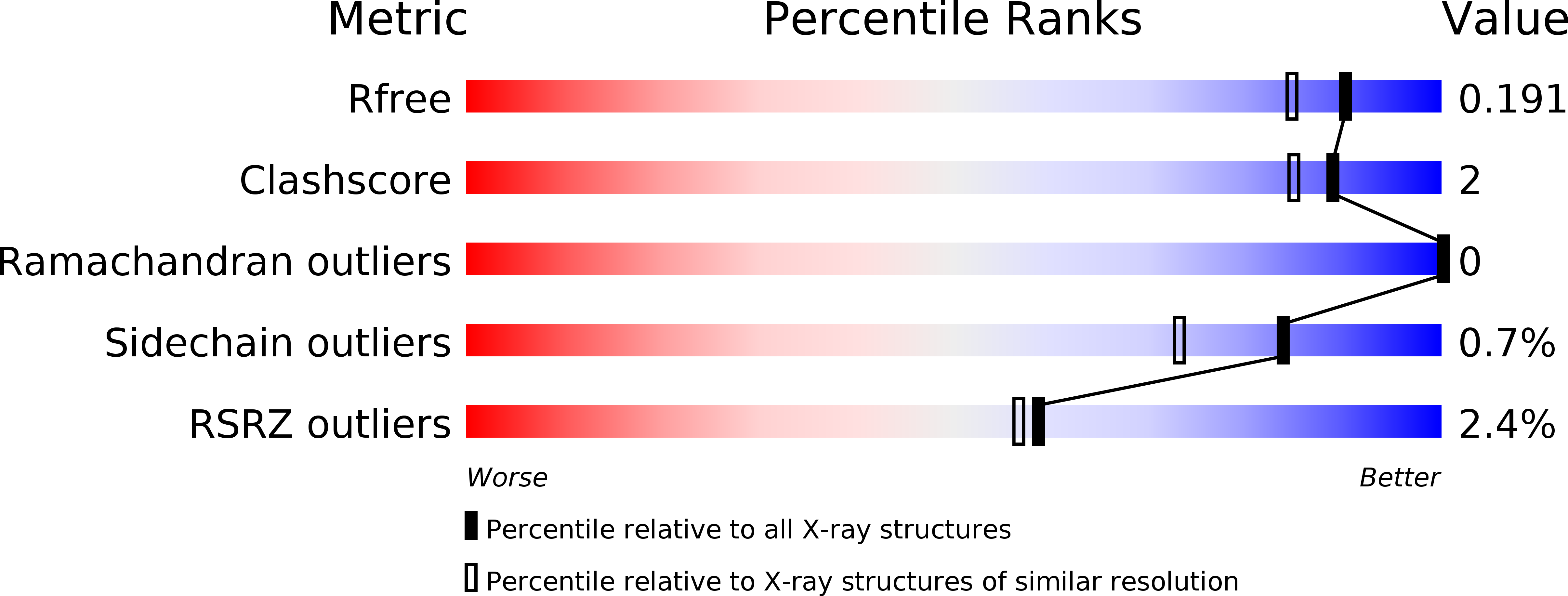
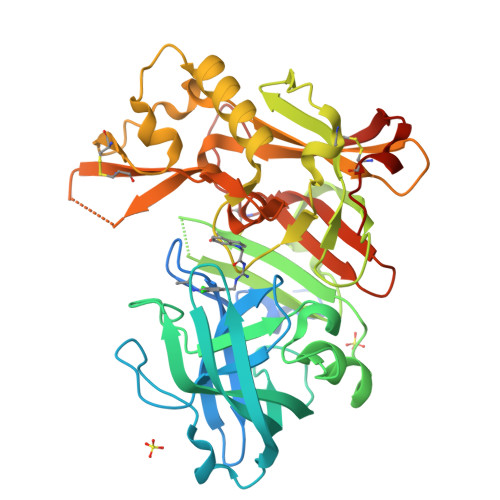
Proteolytic cleavage of amyloid precursor protein by β-secretase (BACE1) is a key step in generating the N-terminal of β-amyloid (Aβ), which further forms into amyloid plaques that are considered as the hallmark of Alzheimer's disease. Inhibitors of BACE1 can reduce the levels of Aβ and thus have a therapeutic potential for treating the disease. We report here the identification of a series of small molecules bearing an indole acylguanidine core structure as potent BACE1 inhibitors. The initial weak fragment was discovered by virtual screening, and followed with a hit-to-lead optimization. With the aid of co-crystal structures of two discovered inhibitors (compounds 19 and 25) with BACE1, we explored the SAR around the indole and aryl groups, and obtained several BACE1 inhibitors about 1,000-fold more potent than the initial fragment hit. Accompanying the lead optimization, a previously under-explored sub-site opposite the flap loop was redefined as a potential binding site for later BACE1 inhibitor design.
Organizational Affiliation:
State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, Zhangjiang Hi-Tech Park, Shanghai 201203, China.