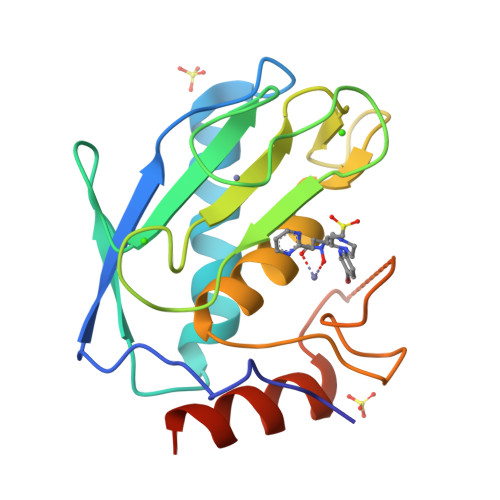
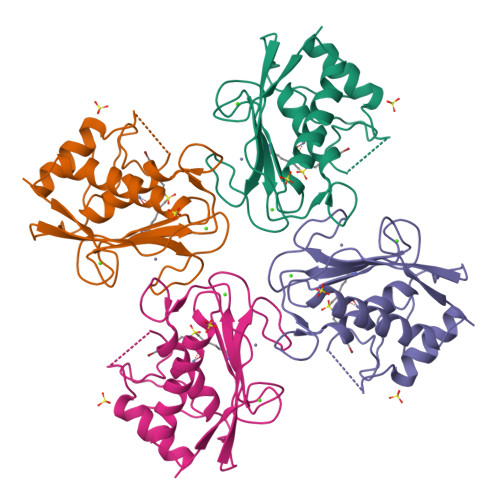
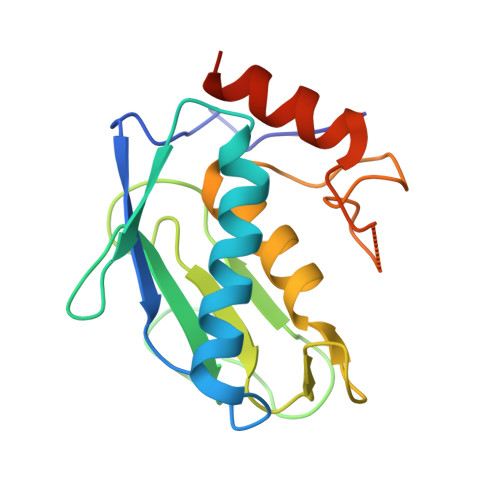
The Discovery of Mmp7 Inhibitors Exploiting a Novel Selectivity Trigger.
Edman, K., Furber, M., Hemsley, P., Johansson, C., Pairaudeau, G., Petersen, J., Stocks, M., Tervo, A., Ward, A., Wells, E., Wissler, L.(2011) ChemMedChem 6: 769
- PubMed: 21520417
- DOI: https://doi.org/10.1002/cmdc.201000550
- Primary Citation of Related Structures:
2Y6C, 2Y6D
Organizational Affiliation:
Global Structural Chemistry, AstraZeneca Mölndal, Mölndal, Sweden. Karl.Edman@astrazeneca.com