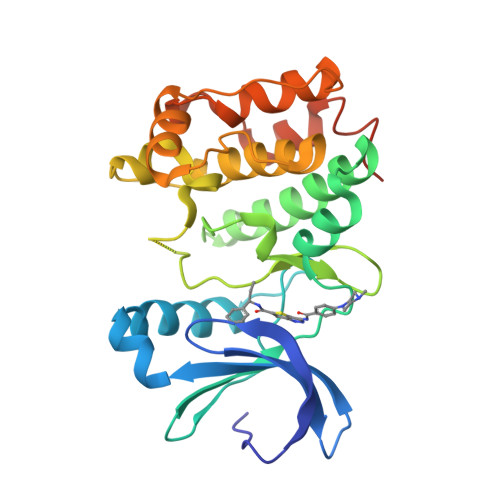
Thieno[3,2-C]Pyrazoles: A Novel Class of Aurora Inhibitors with Favorable Antitumor Activity.
Bindi, S., Fancelli, D., Alli, C., Berta, D., Bertrand, J.A., Cameron, A.D., Cappella, P., Carpinelli, P., Cervi, G., Croci, V., D'Anello, M., Forte, B., Laura Giorgini, M., Marsiglio, A., Moll, J., Pesenti, E., Pittala, V., Pulici, M., Riccardi-Sirtori, F., Roletto, F., Soncini, C., Storici, P., Varasi, M., Volpi, D., Zugnoni, P., Vianello, P.(2010) Bioorg Med Chem 18: 7113
- PubMed: 20817473
- DOI: https://doi.org/10.1016/j.bmc.2010.07.048
- Primary Citation of Related Structures:
2XRU - PubMed Abstract:
A novel series of 3-amino-1H-thieno[3,2-c]pyrazole derivatives demonstrating high potency in inhibiting Aurora kinases was developed. Here we describe the synthesis and a preliminary structure-activity relationship, which led to the discovery of a representative compound (38), which showed low nanomolar inhibitory activity in the anti-proliferation assay and was able to block the cell cycle in HCT-116 cell line. This compound demonstrated favorable pharmacokinetic properties and good efficacy in the HL-60 xenograft tumor model.
Organizational Affiliation:
Nerviano Medical Sciences-Oncology, via Pasteur 10, 20014 Nerviano, Milan, Italy. simona.bindi@nervianoms.com