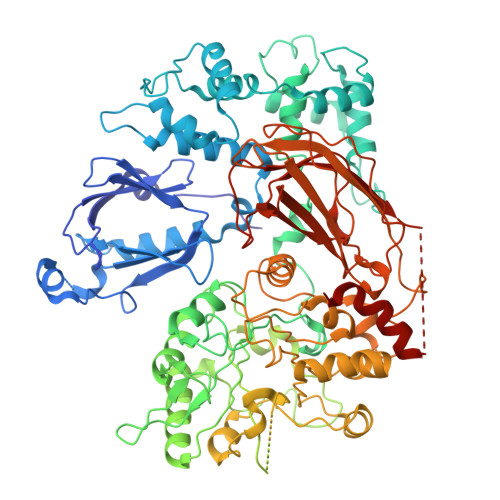
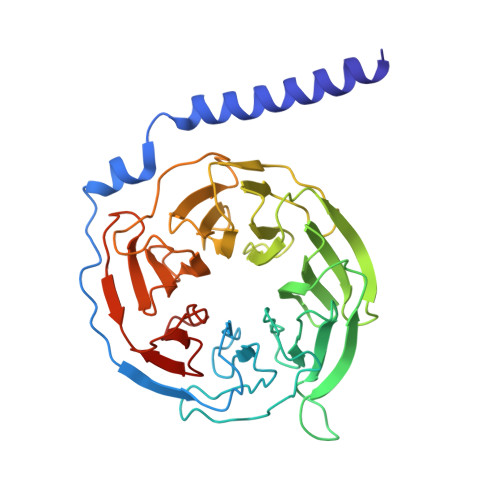
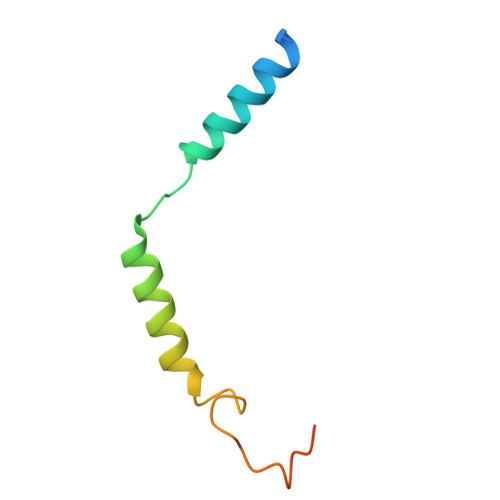
G beta gamma engages PLC beta 3 at multiple sites to reorient and facilitate its activation.
Fisher, I.J., Senarath, K., Outlaw, K., Muralidharan, K., Garland-Kuntz, E.E., Van Camp, M., Komay, T., Inoue, A., Kostenis, E., Lambert, N.A., Lyon, A.M.(2026) bioRxiv
- PubMed: 41648476
- DOI: https://doi.org/10.64898/2026.01.14.699417
- Primary Citation of Related Structures:
9Y7H, 9YAO, 9YAP - PubMed Abstract:
Phospholipase C β (PLCβ) enzymes are activated by heterotrimeric G protein subunits, increasing hydrolysis of phosphatidylinositol-4,5-bisphosphate (PIP2) at the plasma membrane. All four human PLCβ isoforms (PLCβ1-4) are activated by Gα q , while PLCβ1-3 are activated to varying extents by Gβγ. The binding sites for Gα q on PLCβ are well-established and much has been learned about its mechanism of activation, but comparatively little is known about Gβγ-dependent activation. In this work, we used cryo-electron microscopy (cryo-EM) single particle analysis (SPA), functional assays, and bioluminescence resonance energy transfer (BRET) to investigate how Gβγ interacts with PLCβ3 in concert with activated Gα q to regulate phospholipase activity. Gβγ heterodimers bind multiple surfaces of PLCβ3 to promote activation but alone do not recruit the enzyme to the plasma membrane. Instead, Gβγ facilitates activation by Gα q , most likely by reorienting the phospholipase catalytic site at the membrane to maximize PIP2 hydrolysis and downstream Ca 2+ release. Cell-based functional assays demonstrate that Gβγ is required for maximal PLCβ3 activation even when G q heterotrimers are the sole source of Gβγ. Together, these findings demonstrate that Gβγ acts as a critical positive allosteric modulator that regularly acts in concert with Gα q to activate PLCβ3 at the plasma membrane.
- James Tarpo Jr. and Margaret Tarpo Department of Chemistry, Purdue University, West Lafayette, IN, USA.
Organizational Affiliation: