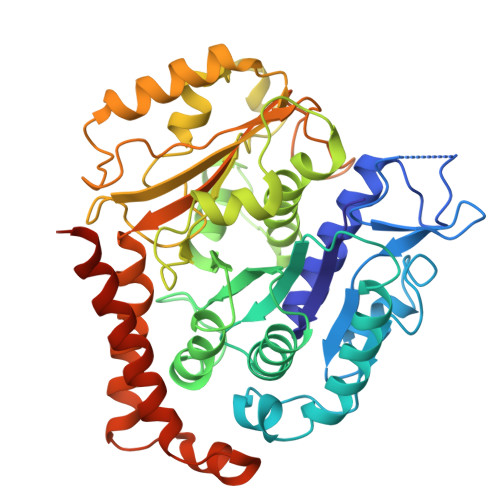
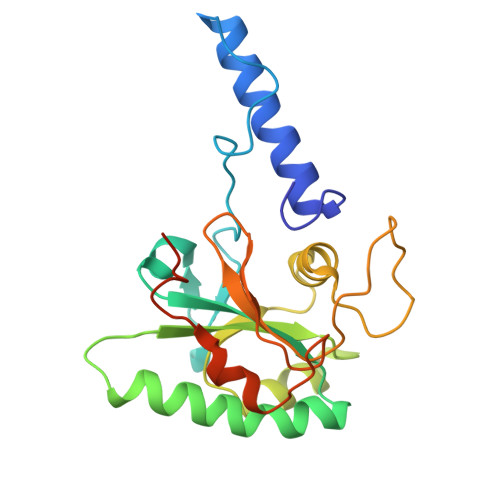
Atomic models of the Toxoplasma cell invasion machinery.
Zeng, J., Fu, Y., Qian, P., Huang, W., Niu, Q., Beatty, W.L., Brown, A., Sibley, L.D., Zhang, R.(2026) Nat Struct Mol Biol 33: 157-170
- PubMed: 41366525
- DOI: https://doi.org/10.1038/s41594-025-01728-w
- Primary Citation of Related Structures:
9Y9Z, 9YA0, 9YA1, 9YA2, 9YA3 - PubMed Abstract:
Apicomplexan parasites, responsible for toxoplasmosis, cryptosporidiosis and malaria, invade host cells through a unique gliding motility mechanism powered by actomyosin motors and a dynamic organelle called the conoid. Here, using cryo-electron microscopy, we determined structures of four essential complexes of the Toxoplasma gondii conoid: the preconoidal P2 ring, tubulin-based conoid fibers, and the subpellicular and intraconoidal microtubules. Our analysis identified 40 distinct conoid proteins, several of which are essential for parasite lytic growth, as revealed through genetic disruption studies. Comparative analysis of the tubulin-containing complexes sheds light on their functional specialization by microtubule-associated proteins, while the structure of the preconoidal ring pinpoints the site of actin polymerization and initial translocation, enhancing our mechanistic understanding of gliding motility and, therefore, parasite invasion.
- Department of Biochemistry, Washington University in St. Louis, School of Medicine, St. Louis, MO, USA.
Organizational Affiliation: