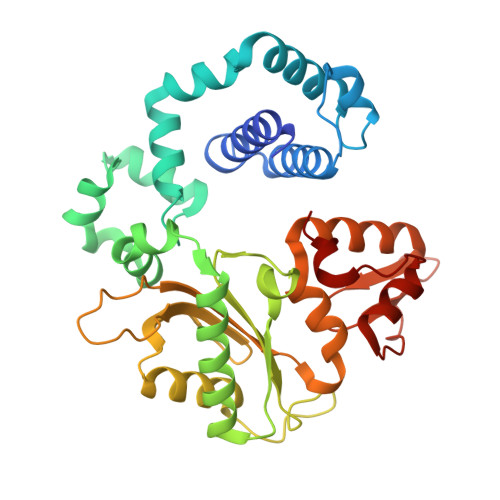
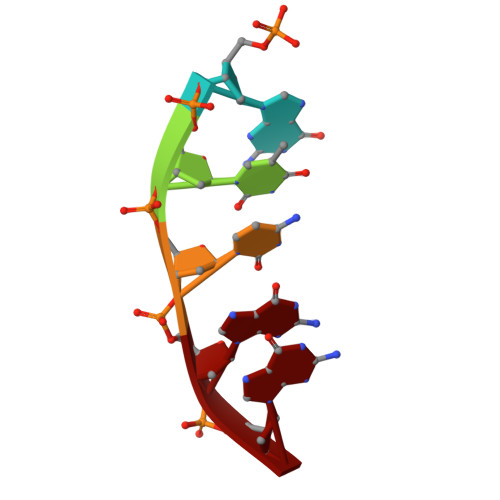
The S180R Human Germline Variant of DNA Polymerase beta Is a Low Fidelity Enzyme with Reduced Flexibility of the Fingers Domain.
Sawyer, D.L., Eckenroth, B.E., Chavira, C., Alnajjar, K., Hanley, J.P., Dragon, J.A., Doublie, S., Sweasy, J.B.(2026) Biochemistry
- PubMed: 41524291
- DOI: https://doi.org/10.1021/acs.biochem.5c00628
- Primary Citation of Related Structures:
9Y15, 9Y16, 9Y17, 9Y18, 9Y19, 9Y1A, 9Y1B, 9Y1C, 9Y1D, 9Y1E, 9Y1F, 9Y1G, 9Y1H, 9Y1I, 9Y1J, 9Y1K - PubMed Abstract:
DNA polymerase β (Pol β) is an important polymerase that functions in DNA repair within the Base Excision Repair and Non-Homologous End-Joining pathways. It is estimated to function in the repair of up to 50,000 DNA lesions per cell per day, within the base excision repair pathway (BER). Given the significant role Pol β plays in repairing DNA, genetic variants of Pol β have the potential to perturb repair, resulting in mutation accumulation which can potentiate cancer formation. Here we identify an unstudied human germline variant of Pol β, the S180R variant (rs1585898410), which introduces a significant amino acid alteration within the dNTP binding pocket of the enzyme. We demonstrate that S180R is a low fidelity variant of Pol β due to its loss of the ability to discriminate correct nucleotides from incorrect nucleotides. We also show that this variant exhibits a much slower rate of nucleotide incorporation, which could further disrupt repair capacity in vivo . Structural data reveal that this variant not only has structural changes that may disrupt dNTP binding but also a loss of primer terminus positioning and dynamic flexibility of the fingers domain in the binary state, which likely are driving the low fidelity of S180R Pol β. This study highlights the importance of binary positioning and nucleotide coordinating residues for maintaining nucleotide selectivity, polymerase function, and fidelity. It also emphasizes the importance of further study of this human germline Pol β variant in vivo .
- Department of Cell and Molecular Medicine, University of Arizona, 1515 N Campbell Avenue, Tucson, Arizona 85724, United States.
Organizational Affiliation: