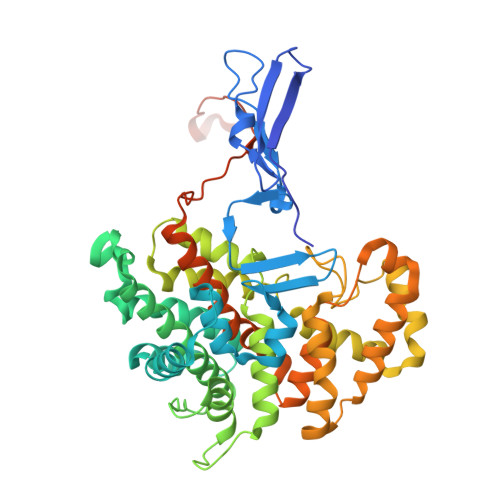
Crystal structure of glyoxysomal citrate synthase 3 from Arabidopsis thaliana reveals a novel oligomeric state.
Nishio, K., Takagi, K., Mizushima, T.(2026) J Struct Biol : 108293-108293
- PubMed: 41580056
- DOI: https://doi.org/10.1016/j.jsb.2026.108293
- Primary Citation of Related Structures:
9X0I, 9XAU, 9XDG - PubMed Abstract:
Citrate synthase (CS) is a pivotal enzyme in carbohydrate and energy metabolism, with distinct isoforms present in various eukaryotic compartments, including mitochondria and glyoxysomes in plants. While CSs exhibit diverse oligomeric states, detailed structural information on higher plant non-mitochondrial Type II CSs has been limited. We herein determined the crystal structures of CS 3 from Arabidopsis thaliana (AtCSY3) in complex with oxaloacetate (OAA) and acetyl-coenzyme A (CoA)-OAA at resolutions of 2.0 and 1.7 Å, respectively. These structures revealed that AtCSY3 can form a homo-tetrameric assembly that is distinct from the hexameric Escherichia coli CS and the octameric Ananas comosus CS. The tetrameric arrangement observed in the crystal structure is mediated by hydrogen-bonding and hydrophobic interactions between subunits. Gel filtration chromatography further suggests the presence of a tetrameric species in solution under the purification conditions. Ligand density was observed near the interface between the two dimers in the tetrameric structure; however, no experimental evidence is currently available to determine whether ligand binding affects the oligomeric state or enzymatic activity of AtCSY3. These structures illustrate the structural diversity of CS oligomerization and provide a structural basis for studies of plant glyoxysomal CSs.
- Department of Science, Graduate School of Science, University of Hyogo, 2167, Shosha, Himeji 671-2280, Japan.
Organizational Affiliation: