Functional and Structural Characterization of a Novel Anti-His-tag Antibody, HisMab-1.
Hitomi, N., Hoshi, S., Kaneko, M.K., Kato, R., Iwasaki, K., Takagi, J., Kato, Y., Harada-Hikita, A., Arimori, T.(2025) J Mol Biology 438: 169574-169574
- PubMed: 41349762
- DOI: https://doi.org/10.1016/j.jmb.2025.169574
- Primary Citation of Related Structures:
9VX2, 9VX3 - PubMed Abstract:
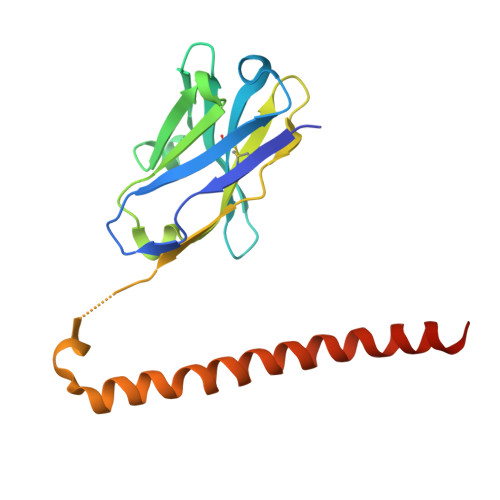
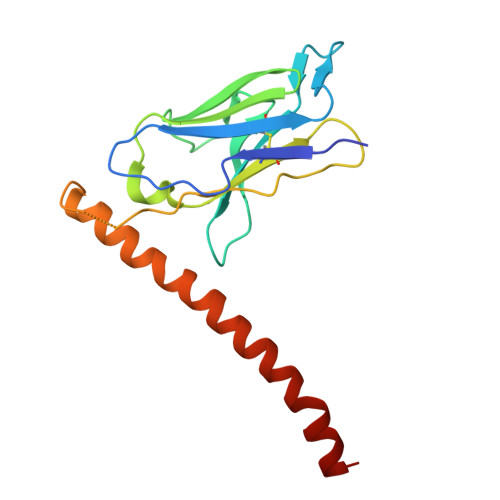
The polyhistidine tag (His-tag) is one of the most widely used peptide tags for the purification of recombinant proteins, owing to its compatibility with immobilized metal affinity chromatography. While numerous anti-His-tag antibodies are commercially available, their quantitative affinity data and structural insights are limited. Here, we present a detailed physicochemical and structural characterization of a novel anti-His-tag antibody, HisMab-1. Isothermal titration calorimetry showed that the Fab fragment of HisMab-1 binds to a hexahistidine peptide in an enthalpy-driven manner, with a dissociation constant (K D ) of ∼30 nM at a neutral pH. The crystal structure of the HisMab-1-hexahistidine peptide complex at 2.39-Å resolution revealed that HisMab-1 primarily recognizes the first, second, fourth, and fifth histidine residues of the peptide through multiple interactions, including hydrogen bonding and π-π stacking, which collectively contribute to the high specificity of the antibody. Notably, HisMab-1 also binds to a His-tag embedded within a conformationally constrained β-hairpin loop without reducing affinity, highlighting its structural adaptability. These findings establish HisMab-1 as a high-affinity, high-specificity, structurally validated anti-His-tag antibody with broad potential in diverse protein engineering and structural biology applications.
- Laboratory for Protein Synthesis and Expression, Institute for Protein Research, The University of Osaka, 3-2, Yamadaoka, Suita, Osaka 565-0871, Japan.
Organizational Affiliation: