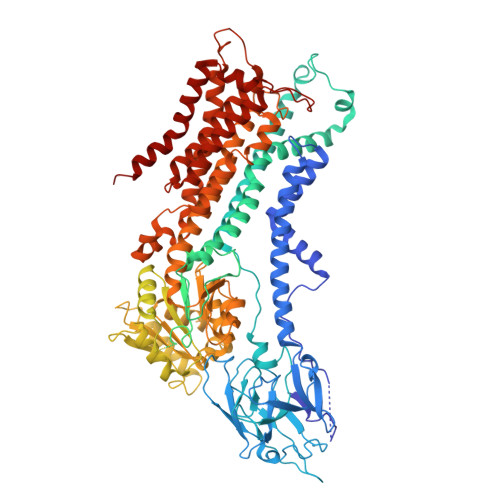
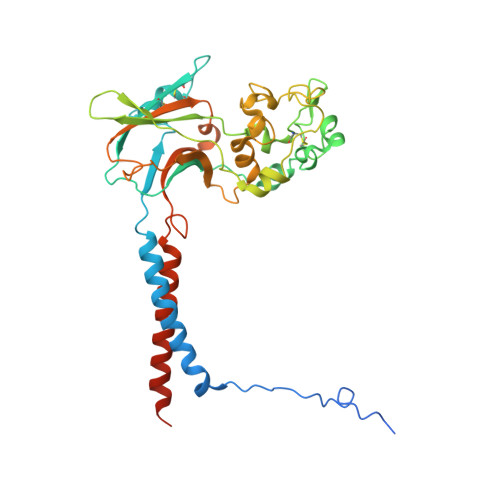
Cryo-EM structure of the ATP11C Q79E mutant reveals the structural basis for altered Phospholipid recognition.
Qian, Y., Gopalasingam, C.C., Gerle, C., Shigematsu, H., Abe, K., Oshima, A.(2025) J Biological Chem 302: 110935-110935
- PubMed: 41237907
- DOI: https://doi.org/10.1016/j.jbc.2025.110935
- Primary Citation of Related Structures:
9VKG, 9VNT, 9VQ2, 9VSL - PubMed Abstract:
Closely related P4-ATPases, ATP11A and ATP11C, act as major phospholipid flippases in the plasma membrane of mammalian cells, with strict substrate specificity for phosphatidylserine (PS) and phosphatidylethanolamine (PE), but not for phosphatidylcholine (PC), thereby contributing to the asymmetric distribution of PS and PE across bilayers. A previously reported disease-associated Q84E mutation in ATP11A confers the ability to flip PC, implicating the involvement of this conserved residue in substrate specificity. We performed cryo-EM analysis for the equivalent mutant Q79E of ATP11C to address the structural basis for its unusual substrate specificity. Measurement of ATPase activity revealed that the ATP11C Q79E mutant retained PS-dependent activity, whilst gaining robust PC-dependent activity, indicative of expanded substrate specificity, consistent with reported properties in ATP11A Q84E. The cryo-EM structure of ATP11C Q79E mutant in the PC-occluded E2-P i state revealed a PC molecule in a reshaped binding pocket. Due to the Q79E mutation and associated conformational changes in its surrounding residues, including Ser91and Asn352, the binding pocket has additional space to accommodate the bulky choline headgroup. Our results provide structural and functional insights into how a single point mutation can alter substrate specificity in a P4-ATPase.
- Department of Chemistry, Faculty of Science, Hokkaido University, Japan; Graduate School of Pharmaceutical Sciences, Nagoya University, Nagoya, Japan.
Organizational Affiliation: