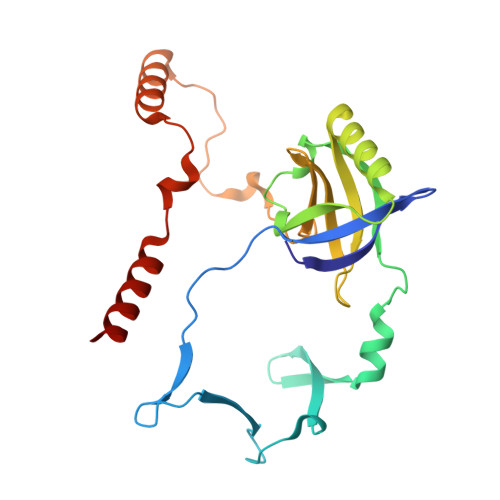
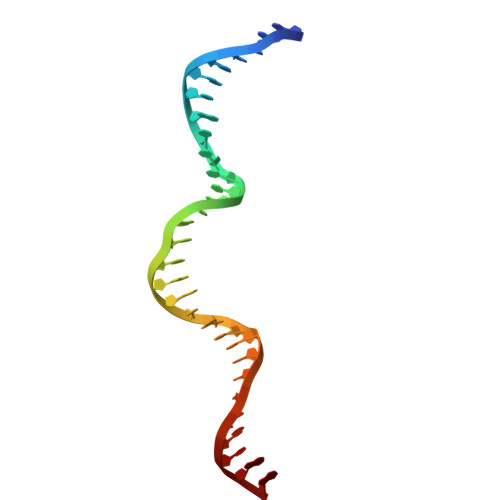
Structure of Bacillus subtilis Ku-mediated DNA synaptic complex.
Kim, W.J., Kim, J., Jo, M., Kim, Y., Kim, M.S.(2026) Nucleic Acids Res 54
- PubMed: 41118517
- DOI: https://doi.org/10.1093/nar/gkaf1036
- Primary Citation of Related Structures:
9VNQ - PubMed Abstract:
DNA double-strand breaks (DSBs) pose a severe threat to genomic integrity, and cells rely on two major pathways for repair: homologous recombination and non-homologous end joining (NHEJ). While eukaryotic NHEJ requires a multi-component assembly including the Ku70/80 heterodimer, bacterial NHEJ operates with a simpler toolkit comprising a Ku homodimer and the multifunctional LigD. Despite this simplicity, the mechanism by which broken DNA ends are bridged together has remained unclear in bacterial NHEJ. Here, we present a cryo-electron microscopy structure of the Bacillus subtilis Ku (bsKu)-DNA complex at 2.74 Å resolution, capturing two blunt DNA ends bridged by a Ku protein alone. Supported by further biochemical assays, we propose an integrated model in which oligomeric arrays of Ku homodimers bridge and stabilize two DNA ends, facilitating efficient DSB repair in Bacillus subtilis. This work reveals a bsKu-mediated DNA bridging mechanism distinct from the eukaryotic system and provides critical structural insight into prokaryotic DNA repair.
- Department of Life Sciences, Pohang University of Science and Technology, Pohang, Gyeongbuk, 37673, Republic of Korea.
Organizational Affiliation: