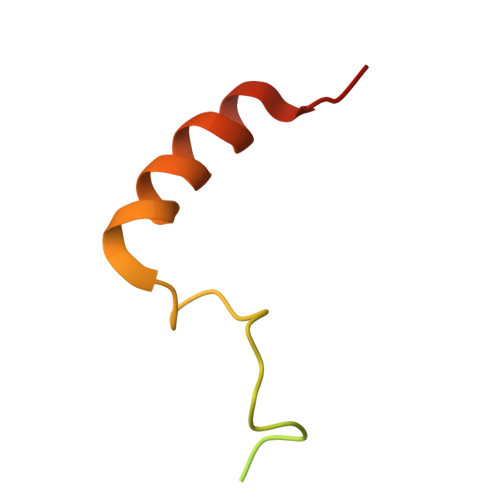
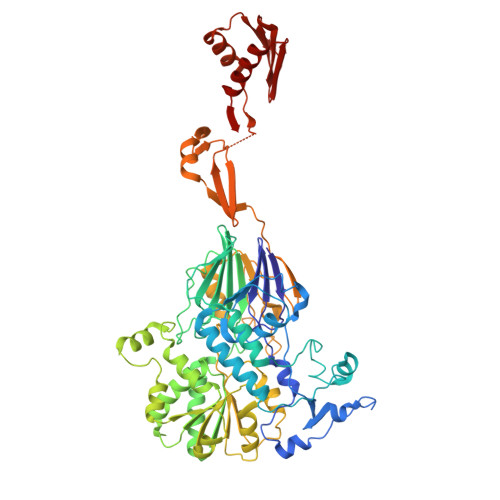
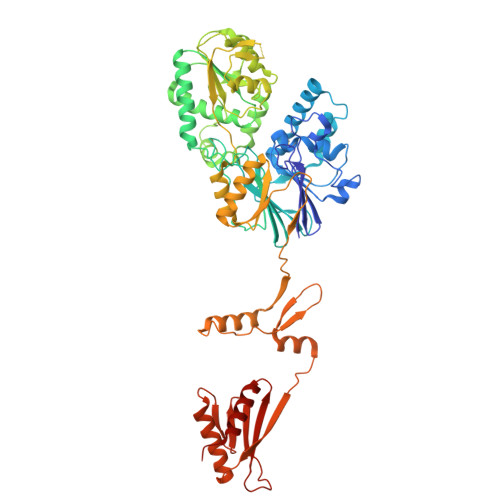
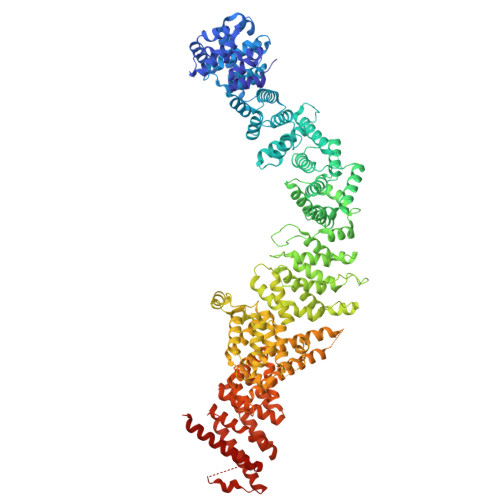
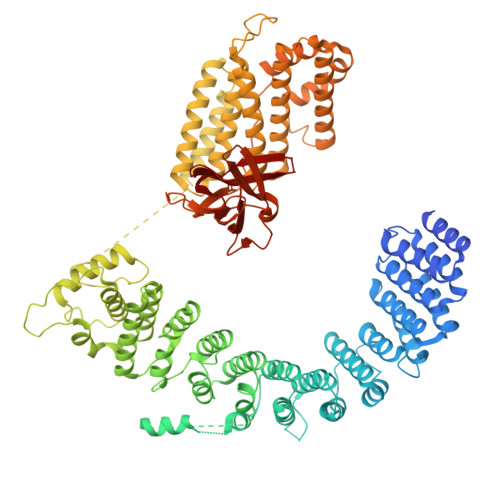
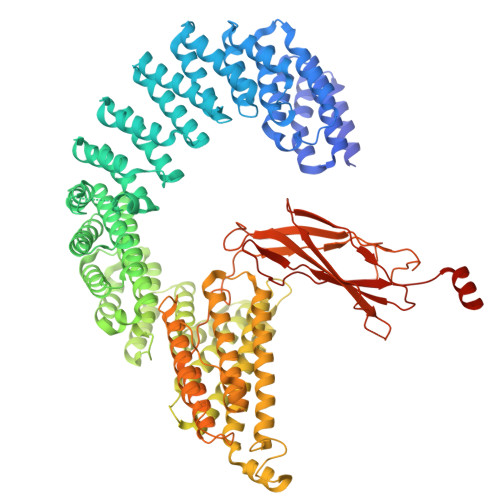
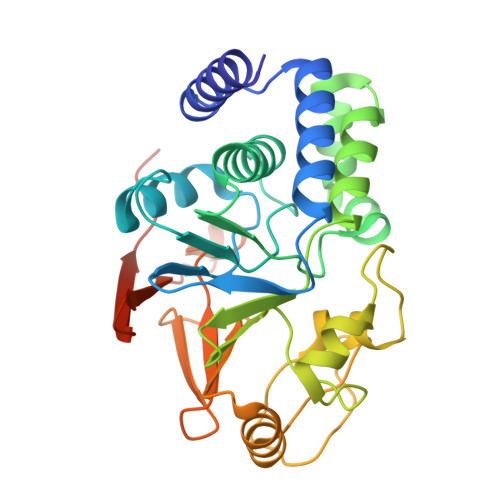
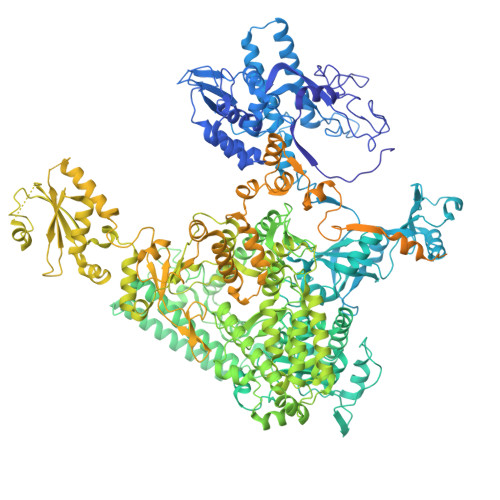
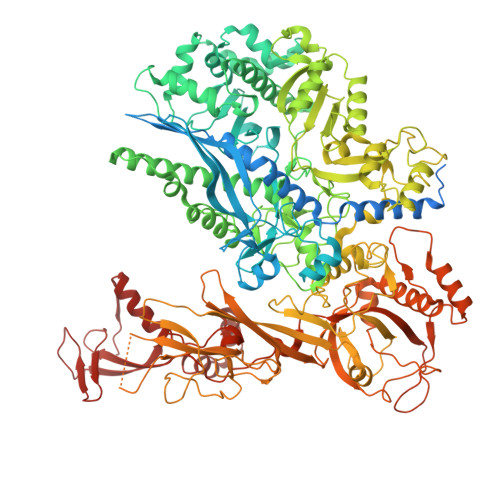
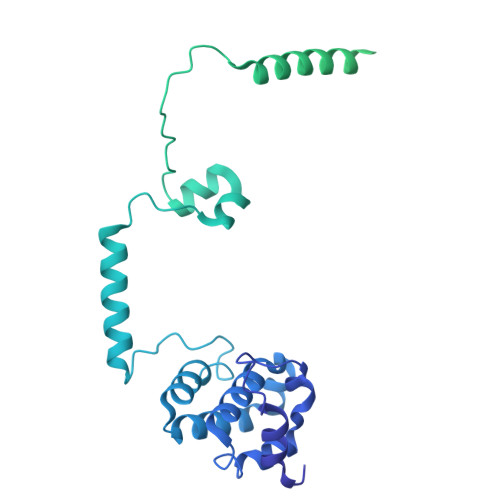
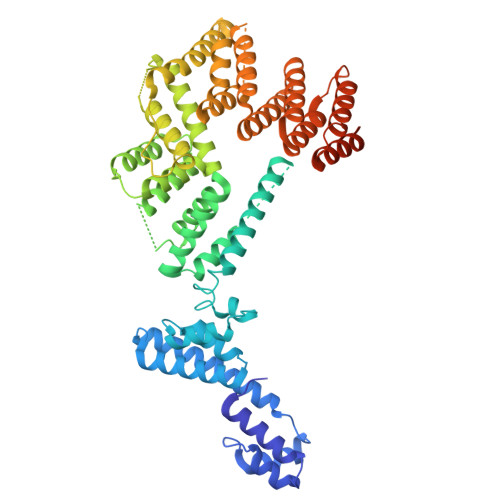
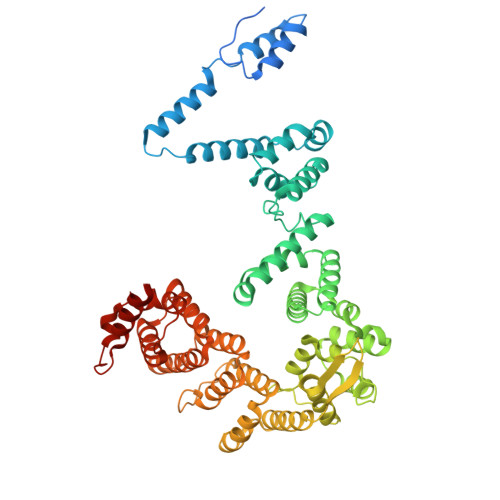
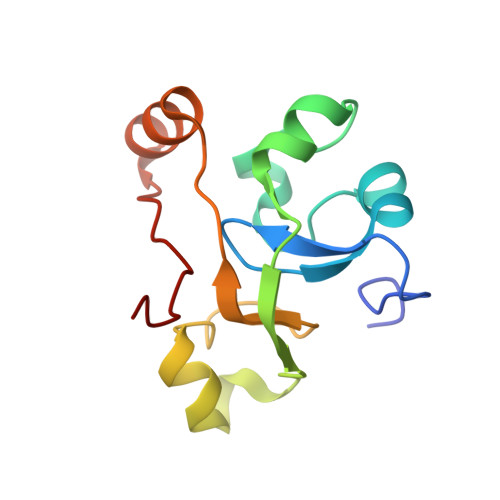
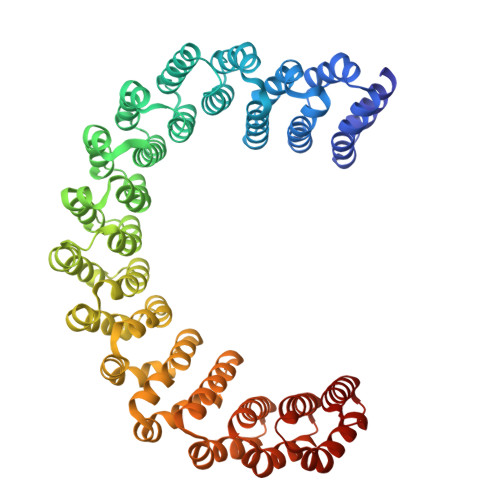DSS1 is required for proper Integrator-PP2A function.
Xu, C., Zhou, Q.X., Zheng, H., Song, A., Zhao, W.Y., Xu, T.T., Xiong, Y., Zhang, Y.J., Huang, Z., Xu, Y., Cheng, J., Chen, F.X.(2025) Nat Commun 16: 6206-6206
- PubMed: 40617815
- DOI: https://doi.org/10.1038/s41467-025-61257-4
- Primary Citation of Related Structures:
9VD9 - PubMed Abstract:
Integrator-PP2A (INTAC) is a highly modular complex orchestrating the transition of paused RNA polymerase II into productive elongation or promoter-proximal premature termination, with its loss resulting in transcription dysregulation and genome instability. Here, we identify human DSS1-a flexible 70-residue protein found in multiple functionally diverse complexes including the 26S proteasome-as an integral subunit of the INTAC backbone. Structural analysis of DSS1-INTAC, both alone and in association with paused polymerase, demonstrates intimate interactions between DSS1 and the INTAC backbone. We identify tryptophan 39 of DSS1 as being critical for interacting with INTAC and find that its mutation disrupts DSS1's interaction with INTAC, while maintaining DSS1's interaction with the proteasome. This substitution not only impairs INTAC-dependent transcriptional regulation, but also reveals that INTAC is DSS1's major chromatin-bound form. Together, our findings reveal a role for DSS1 in supporting the structure and regulatory functions of INTAC.
- Shanghai Key Laboratory of Anesthesiology and Brain Functional Modulation, Clinical Research Center for Anesthesiology and Perioperative Medicine, Translational Research Institute of Brain and Brain-Like Intelligence, Shanghai Fourth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai, China. conglingxu@tongji.edu.cn.
Organizational Affiliation: