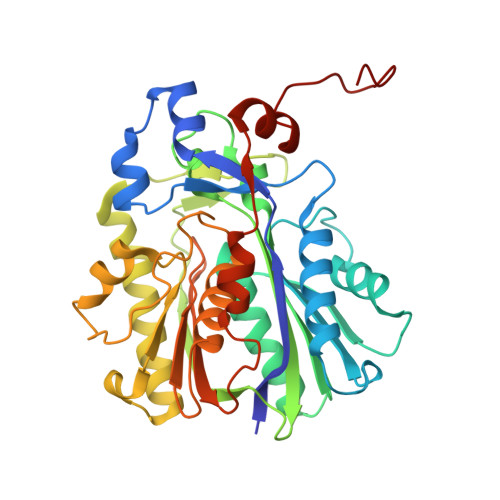
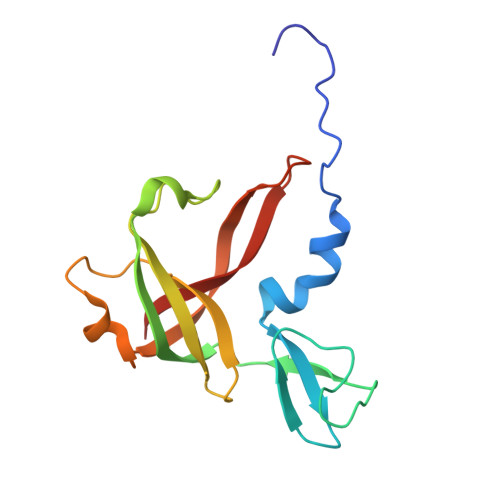
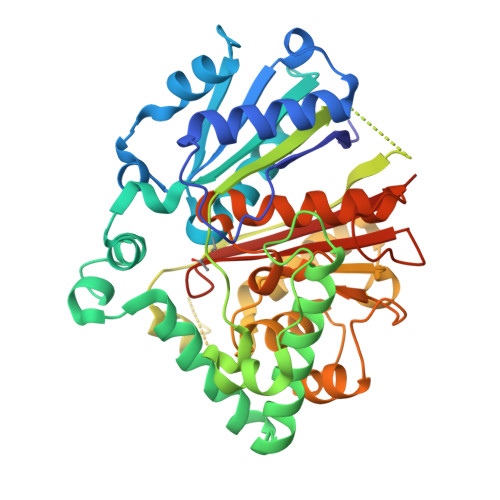
Evolutionary repurposing of a metabolic thiolase complex enables antibiotic biosynthesis.
Liao, G., Sun, R., Shen, Z., Luo, Z., Pang, C., Shen, Z., Wei, A., Mi, C., Wu, G., Li, F., Li, Y.X., Hoi, K.K., Pan, X., Tang, X.(2026) Nat Commun
- PubMed: 41617701
- DOI: https://doi.org/10.1038/s41467-026-68910-6
- Primary Citation of Related Structures:
9VBO, 9VBT - PubMed Abstract:
The functional diversification of biosynthetic enzymes underlies the chemical richness of natural products, yet how primary metabolic enzymes evolve to acquire specialized functions in secondary metabolism remains elusive. Here, we report a tripartite enzyme complex from oral Streptococcus species-comprising 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase (HMGS), acetyl-CoA acetyltransferase (ACAT), and a DUF35 protein-that catalyzes an unusual Friedel-Crafts C-acetylation on a pyrrolidine-2,4-dione scaffold, completing the biosynthesis of the antibiotic reutericyclin A. Cryo-electron microscopy of the S. macacae-derived thiolase complex (SmaATase) reveals a conserved architecture resembling the archaeal HMGS/ACAT/DUF35 complex involved in the mevalonate pathway, yet with key catalytic residues rewired to reprogram substrate specificity. Biochemical characterization, molecular modeling, and evolutionary analysis confirmed that the ancestral activity of HMG-CoA synthesis has been lost, while the complex has been repurposed to mediate Friedel-Crafts C-acylation of small molecule acceptors. These findings reveal a rare example of thiolase complex neofunctionalization, shedding light on an underexplored trajectory in enzyme evolution and offering a template for engineering C-C bond-forming catalysts in synthetic biology.
- Institute of Chemical Biology, Shenzhen Bay Laboratory, Shenzhen, China.
Organizational Affiliation: