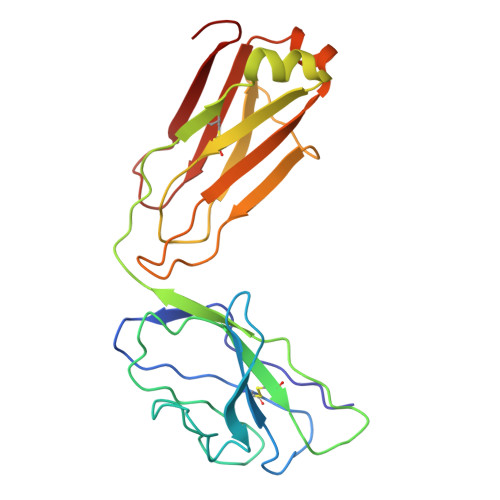
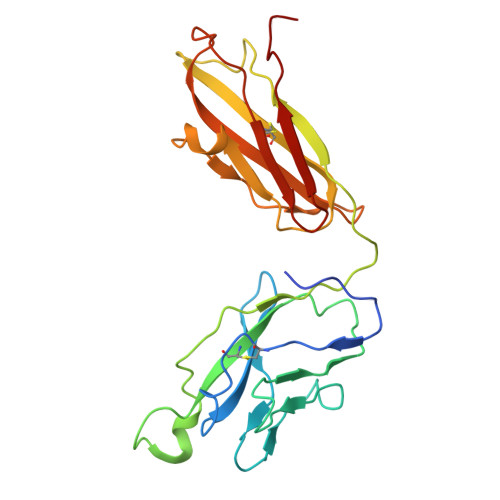
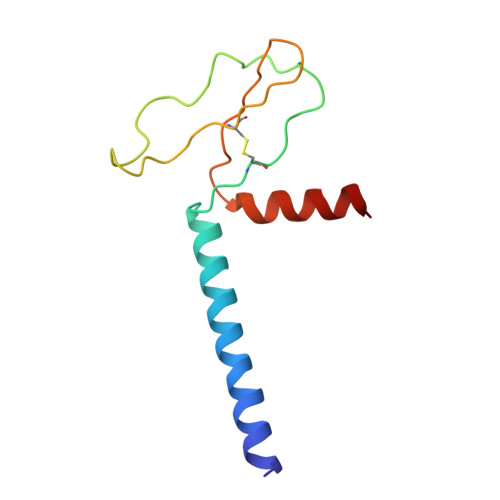
Characterisation of plasmablast-derived HBsAg-specific antibody and its structural basis for binding to native HBsAg dimer.
Ju, B., Liu, Z., Yan, H., Liu, Y., Zhang, L., Ge, X., Wang, X., Si, Z., Zhou, B., Fan, Q., Wang, M., Li, Y., Lai, W., Gan, J., Wang, H., Zhao, J., Xia, Y., Liao, M., Zhang, Z.(2026) Gut
- PubMed: 41663152
- DOI: https://doi.org/10.1136/gutjnl-2025-336641
- Primary Citation of Related Structures:
9UGO - PubMed Abstract:
Plasmablast-derived HBV surface antigen (HBsAg)-specific monoclonal antibody (mAb) and structural basis for binding to native HBsAg are poorly known. We aimed to identify plasmablast-derived HBsAg-specific mAbs, evaluate their antiviral activities and resolve their structure for binding to native HBsAg. A previously vaccinated volunteer was enrolled in this study, who was boosted with a dose of recombinant hepatitis B vaccine and donated the blood sample. Activated plasmablasts were sorted from fresh peripheral blood mononuclear cells and mAbs were expressed. Their gene features, cross-genotypic binding activities and antiviral functions in vitro and in vivo were comprehensively analysed. The cryo-electron microscopy (cryo-EM) was used to determine the structure of representative mAb bound to the native HBsAg. In this study, we cloned a series of HBsAg-specific mAbs directly from clonally expanded plasmablasts from a vaccinated individual. Most of the mAbs displayed cross-reactivities of binding to different genotype HBsAg proteins and antiviral functions such as neutralisation and antibody-dependent cellular phagocytosis. These human anti-HBsAg mAbs, especially SY-4-class and SY-23-class, could be good candidates for antibody drugs. The cryo-EM structure of SY-23 bound to the dimeric HBsAg was determined, revealing its binding mechanism and unprecedented structural detail of the major antigenic loop (AGL) of HBsAg. Overall, our work has uncovered the diverse gene features and varied anti-HBV activities of plasmablast-derived mAbs, providing a series of antibody drug candidates and the long-sought-after atomic model of AGL has paved the way for a wholistic characterisation of the AGL's dynamic conformation during HBV infection and immune response.
- Institute for Hepatology, National Clinical Research Center for Infectious Disease, Shenzhen Third People's Hospital, Department of Biochemistry, the Second Affiliated Hospital, School of Medicine, Southern University of Science and Technology, Shenzhen, Guangdong, China zhangzheng1975@aliyun.com jubin2013@163.com liaomf@sustech.edu.cn yuchenxia@whu.edu.cn.
Organizational Affiliation: