cryo-EM structure of M1 muscarinic acetylcholine receptor-alpha5 helix of G11 protein complex bound to iperoxo and nanobody Nb1B4
Zhang, X., Gao, K., Liu, X.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| M1 muscarinic acetylcholine receptor, de novo design protein | 435 | Homo sapiens | Mutation(s): 0 | 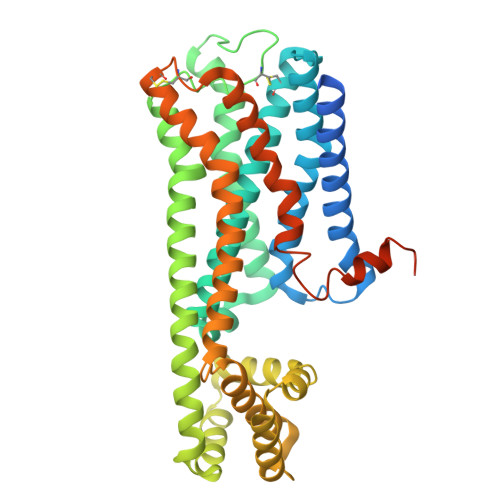 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein subunit alpha-11 | 27 | Homo sapiens | Mutation(s): 0 Gene Names: GNA11, GA11 EC: 3.6.5 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P29992 (Homo sapiens) Explore P29992 Go to UniProtKB: P29992 | |||||
PHAROS: P29992 GTEx: ENSG00000088256 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P29992 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody Nb1B4 | 122 | Vicugna pacos | Mutation(s): 0 | 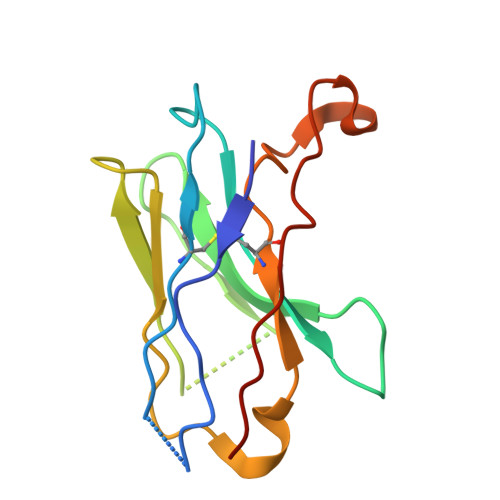 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| IXO (Subject of Investigation/LOI) Query on IXO | D [auth A] | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium C10 H17 N2 O2 WXXOCGISBCTWPW-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32122041 |