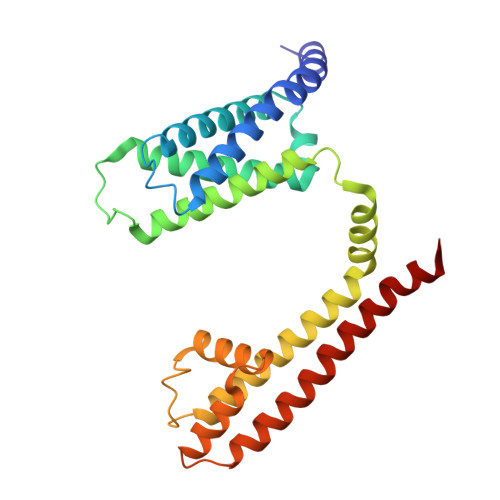
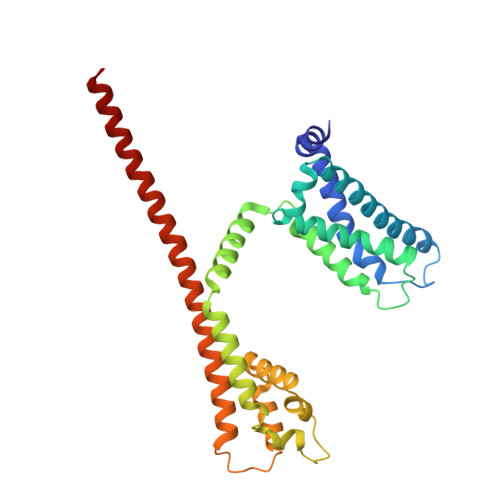
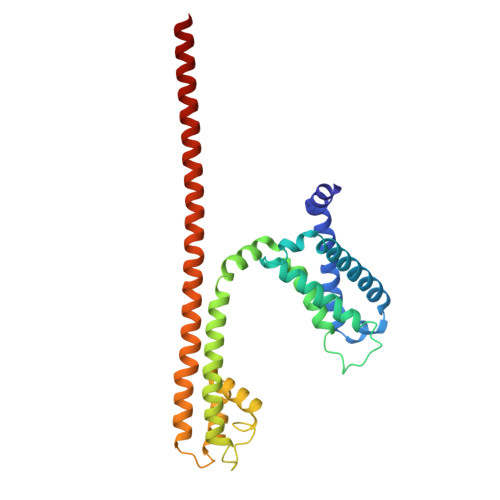
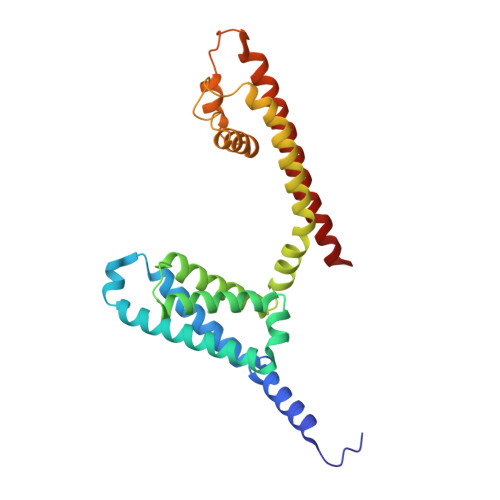
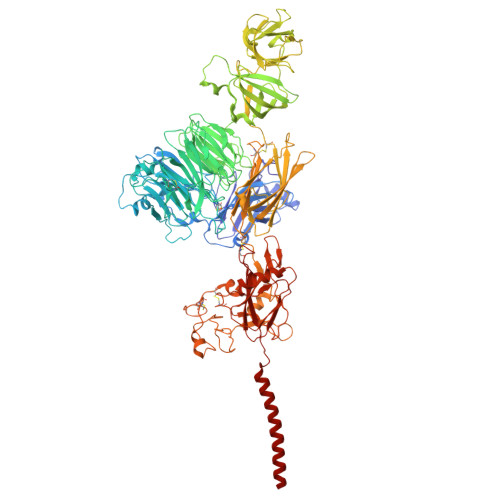
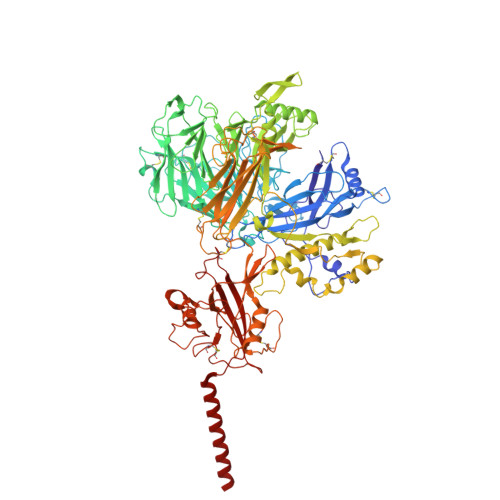
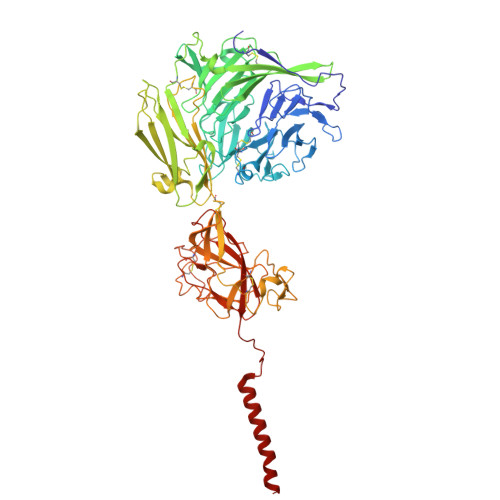
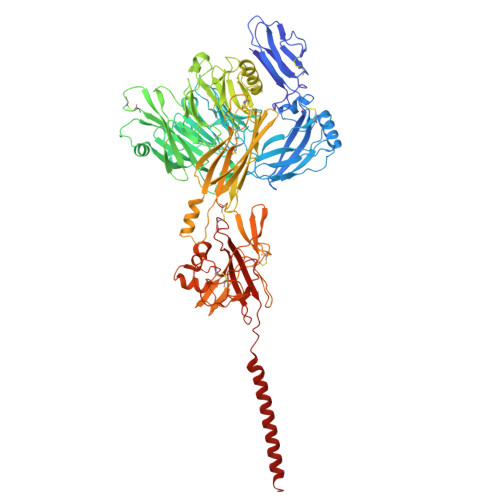
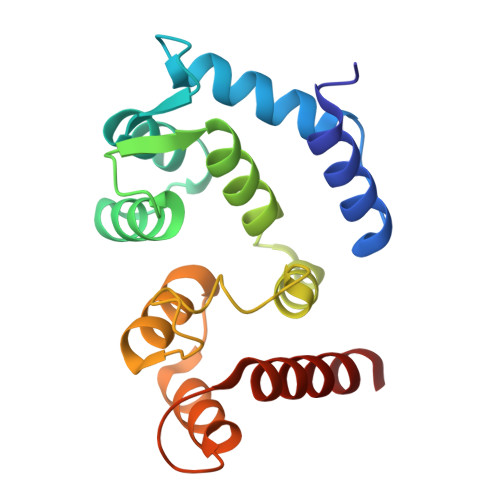
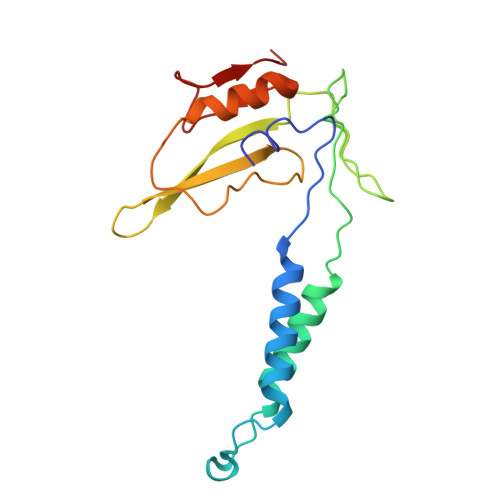
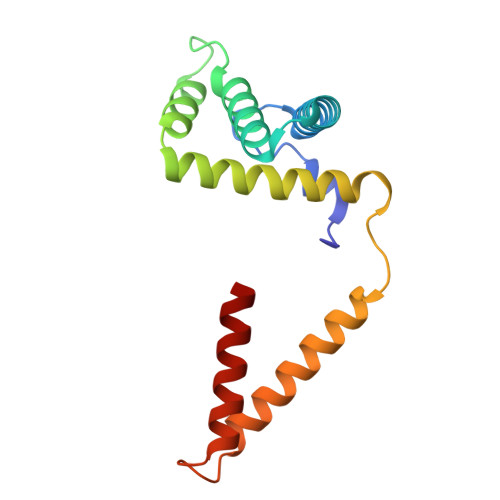
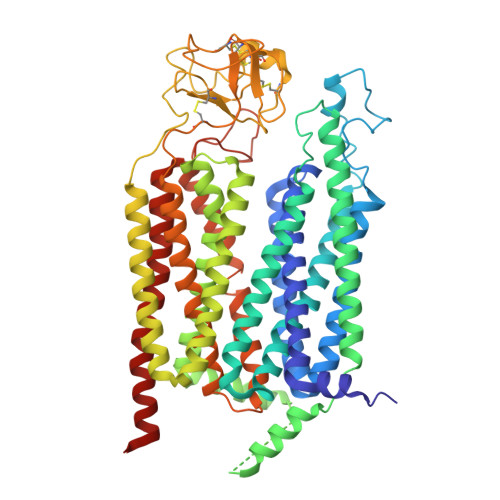
Molecular basis of the higher-order assembly of CatSper.
Xu, Q., Lin, S., Zhao, Q., Ru, Y., Kang, H., Zeng, X., Jiang, M., Yan, Z., Wu, J.(2026) Proc Natl Acad Sci U S A 123: e2510754123-e2510754123
- PubMed: 41490491
- DOI: https://doi.org/10.1073/pnas.2510754123
- Primary Citation of Related Structures:
9UBW, 9UBX - PubMed Abstract:
CatSper serves as the primary Ca 2+ entry pathway in the principal piece of sperm flagellum and is crucial for sperm motility and fertility. Sperm lacking functional CatSper channels fail to undergo hyperactivation during fertilization, leading to complete male infertility. Along the longitudinal axis of the sperm flagellum, staggered CatSper complexes align in a hand-in-hand arrangement, forming a distinctive quadrilinear zigzag arrays known as CatSper nanodomains. However, the molecular details of how CatSper oligomerizes to form such higher-order assembly remain unclear. In this study, we present the cryoelectron microscopy (cryo-EM) structures of native CatSper dimer (~1.5 MDa) and trimer (~2.3 MDa) megacomplexes, which represent the fundamental units of the zigzag array. We reveal the overall configuration of the zigzag assembly by characterizing the two distinct dimer interfaces that mediate CatSper oligomerization. Specifically, we elucidate the interaction details of two extracellular interfaces formed by CATSPERβ and CATSPERγ at the two dimer interfaces, respectively, and find that CATSPERη, a recently identified component of CatSper, constitutes the transmembrane interface within one of these dimer interfaces. Functional studies in mice demonstrate that CATSPERη is essential for the formation of functional CatSper on the sperm flagellum, and CATSPERη-deficient sperm fail to hyperactivate during fertilization, resulting in male infertility both in vivo and in vitro. Our data provide insights into the higher-order assembly of CatSper at the molecular level, offering clues for the development of male contraceptives.
- State Key Laboratory of Gene Expression, School of Life Sciences, Westlake University, Hangzhou, Zhejiang 310024, China.
Organizational Affiliation: