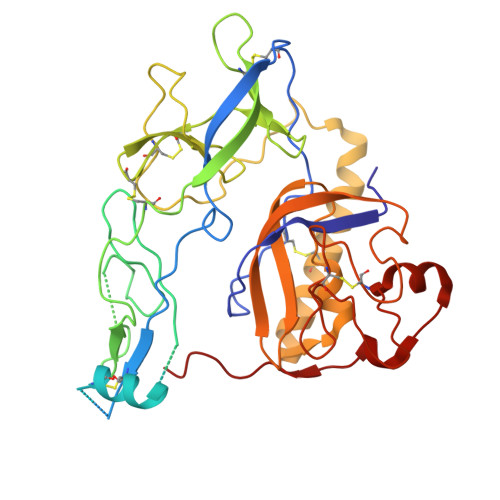
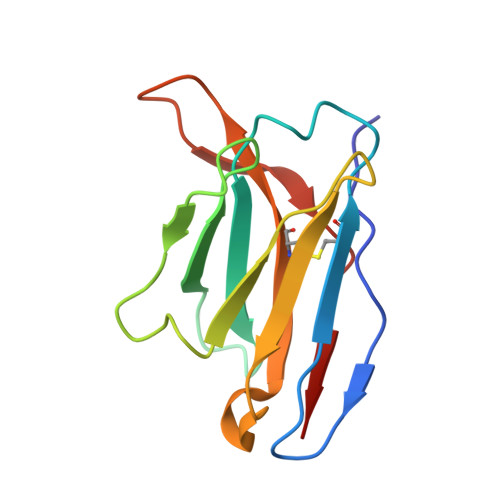
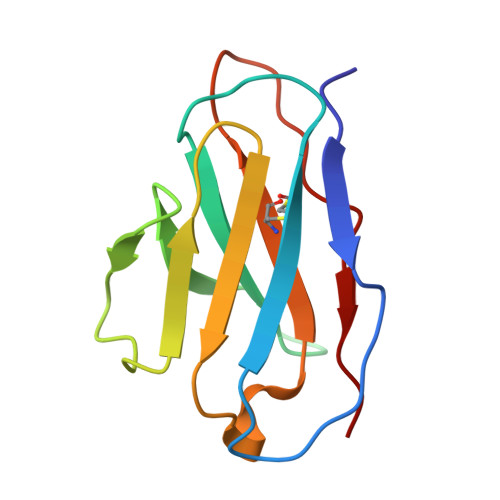
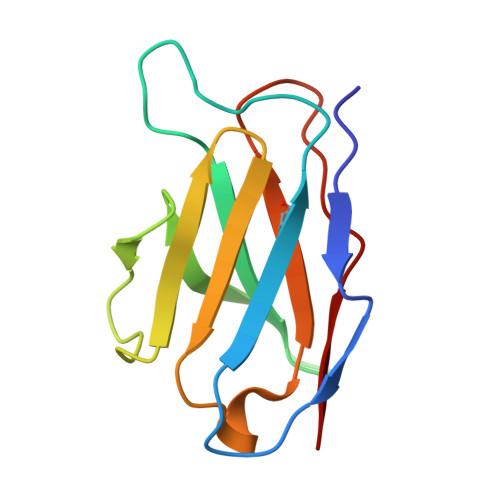
Structural basis for broad neutralization of rabies virus by an antibody cocktail SYN023.
Cao, L., Qu, J., Zhang, C., Chen, R., Wang, J., Fehlner-Gardiner, C., Niezgoda, M., Satheshkumar, P.S., Wang, X., Tsao, E.(2025) Emerg Microbes Infect 14: 2547724-2547724
- PubMed: 40827441
- DOI: https://doi.org/10.1080/22221751.2025.2547724
- Primary Citation of Related Structures:
9UA5 - National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, People's Republic of China.
Organizational Affiliation: