Molecular basis of mitogen-activated protein kinase ERK2 activation by its upstream kinase MEK1
von Velsen, J., Juyoux, P., Piasentin, N., Fisher, H., Lapouge, K., Vadas, O., Gervasio, F.L., Bowler, M.W.(2026) bioRxiv
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
(2026) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dual specificity mitogen-activated protein kinase kinase 1,Uncharacterized protein,Dual specificity mitogen-activated protein kinase kinase 1 | 405 | Homo sapiens, Toxoplasma gondii ME49 | Mutation(s): 2 Gene Names: MAP2K1, MEK1, PRKMK1, TGME49_230180 EC: 2.7.12.2 | 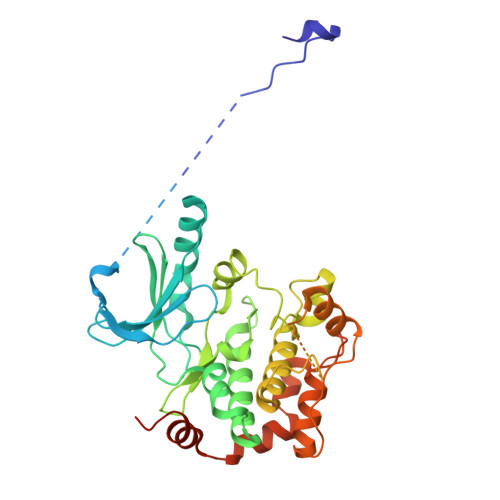 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q02750 (Homo sapiens) Explore Q02750 Go to UniProtKB: Q02750 | |||||
PHAROS: Q02750 GTEx: ENSG00000169032 | |||||
Find proteins for A0A125YG37 (Toxoplasma gondii (strain ATCC 50611 / Me49)) Explore A0A125YG37 Go to UniProtKB: A0A125YG37 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q02750A0A125YG37 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitogen-activated protein kinase 1 | 363 | Homo sapiens | Mutation(s): 1 Gene Names: MAPK1, ERK2, PRKM1, PRKM2 EC: 2.7.11.24 | 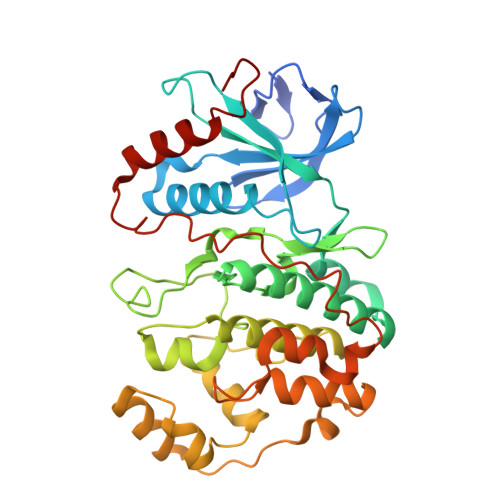 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P28482 (Homo sapiens) Explore P28482 Go to UniProtKB: P28482 | |||||
PHAROS: P28482 GTEx: ENSG00000100030 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P28482 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP (Subject of Investigation/LOI) Query on ADP | C [auth A], E [auth B] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | D [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.2_5419+SVN |
| RECONSTRUCTION | cryoSPARC | v4.7.0 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |