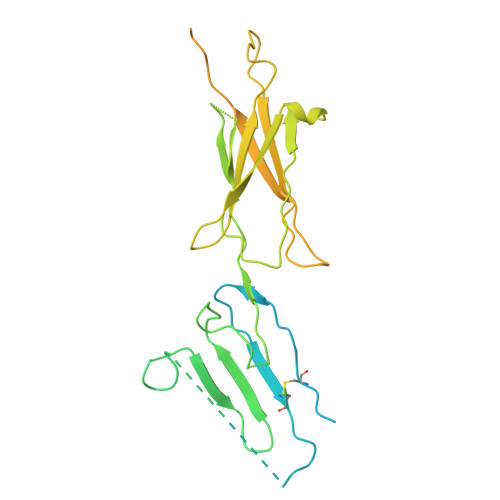
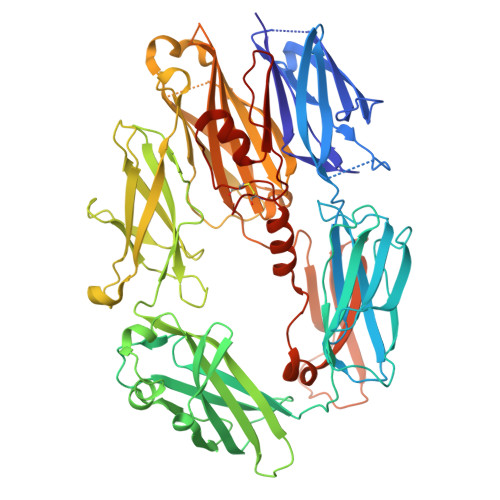
Unveiling the unique interaction mechanism of herpes simplex virus 2 glycoprotein C with C3b.
Rojas Rechy, M.H., Atanasiu, D., Hook, L.M., Cairns, T.M., Saw, W.T., Cahill, A., Guo, Z., Calabrese, A.N., Ranson, N.A., Friedman, H.M., Cohen, G.H., Fontana, J.(2025) bioRxiv
- PubMed: 41256425
- DOI: https://doi.org/10.1101/2025.10.04.680448
- Primary Citation of Related Structures:
9SV8 - PubMed Abstract:
The complement cascade is part of the first line of defence against viral infections, and many viruses have evolved to block it. For example, glycoprotein C (gC) from Herpes Simplex Virus 1 and 2 (gC1 and gC2) facilitates infection by modulating the complement cascade through an interaction with C3b. gC is also involved in attachment and other viral processes. However, our understanding of the molecular mechanisms of gC have been limited due to the absence of a structure. AlphaFold predicts that gC contains a disordered N-terminus and three immunoglobulin-like domains. Here, we generated various gC2 constructs and demonstrated that gC2 domains 1 and 2 are necessary and sufficient to interact with C3b and block the alternative pathway. A gC2 construct lacking the N-terminus in complex with C3b was characterised by cryo-EM at 3.6 Å, providing the first structure for gC2, and revealing that the interaction is predominantly driven by gC2 domain 2 and the MG8 domain of C3b. This structure was confirmed by cross-linking mass spectrometry and by using C3b-blocking antibodies that recognised gC2 linear epitopes at the interface with C3b. Overall, the gC-C3b interaction is different from other C3b-interacting partners, providing a novel mechanism to regulate the complement cascade.
- School of Molecular and Cellular Biology, Faculty of Biological Sciences and Astbury Centre for Structural and Molecular Biology, University of Leeds, Leeds, United Kingdom.
Organizational Affiliation: